Optimizing Caco-2 Permeability with Molecular Pair Analysis: A Guide for Predictive ADMET Profiling
This article provides a comprehensive guide for researchers and drug development professionals on leveraging Matched Molecular Pair Analysis (MMPA) to optimize intestinal permeability predictions using the Caco-2 cell model.
Optimizing Caco-2 Permeability with Molecular Pair Analysis: A Guide for Predictive ADMET Profiling
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on leveraging Matched Molecular Pair Analysis (MMPA) to optimize intestinal permeability predictions using the Caco-2 cell model. It covers the foundational principles of Caco-2 assays and MMPA, details methodological steps for application, addresses common troubleshooting and optimization challenges, and validates the approach through comparisons with machine learning models and industrial case studies. By synthesizing traditional experimental data with modern in silico strategies, this resource aims to enhance the efficiency and accuracy of oral drug candidate optimization, offering practical insights for improving predictive performance in early-stage discovery.
Caco-2 Permeability and Molecular Pairs: Building the Foundation for Predictive ADMET
The Caco-2 cell line, derived from human colorectal adenocarcinoma, has stood the gold standard for in vitro prediction of intestinal drug absorption and permeability for decades [1] [2] [3]. When cultured under specific conditions, these cells spontaneously differentiate into enterocyte-like cells, forming polarized monolayers with tight junctions and well-developed microvilli that mimic the intestinal epithelial barrier [1] [4]. This model's predictive power for passive drug permeability, reproducibility, and relative ease of use has made it indispensable to pharmaceutical research [2]. However, researchers must navigate significant limitations and technical challenges to generate reliable, physiologically relevant data.
The model's relevance stems from its ability to express many morphological and functional characteristics of small intestinal enterocytes despite its colonic origin [4]. Differentiated Caco-2 cells exhibit digestive enzymes, membrane peptidases, disaccharidases, and various uptake and efflux transporters critical for nutrient and drug absorption [1] [4]. Nevertheless, key differences exist between this immortalized cell line and the human small intestine in vivo, particularly regarding transporter expression patterns, metabolic capabilities, and paracellular tightness [1] [2]. Understanding these nuances is fundamental for optimizing permeability studies and properly interpreting results.
Troubleshooting Common Caco-2 Experimental Challenges
Cell Culture and Handling Issues
Q: My Caco-2 cells are taking too long to adhere and grow. What could be wrong?
A: Slow adhesion and growth are inherent traits of Caco-2 cells but can be exacerbated by suboptimal conditions. Key considerations include:
- Serum Concentration: Ensure fetal bovine serum (FBS) concentration is at 20%; reduced serum can impair adhesion [5].
- Medium Composition: Using MEM without non-essential amino acids (NEAA) can decrease growth rate and increase floating cells [5].
- Medium Alkalinity: Check if culture medium appears purple-red (alkaline), which hinders cell adhesion [5].
- Passage Practice: Subculturing at 50% confluence rather than 80% can promote more homogeneous, polarized monolayers [3].
- Digestion Efficiency: Incomplete trypsinization can affect subsequent adhesion; Caco-2 cells typically require 5-10 minutes for digestion and often detach in clusters rather than as single cells [5].
Q: I observe many floating cells and large vacuoles in my cultures. Is this normal?
A: Some floating bright cells and vacuoles are normal characteristics of Caco-2 cells [5]. However, if floating cells become increasingly severe or form large clusters, check for:
- Mycoplasma contamination
- Overly dense feeder layers [3]
- Variations in culture medium composition or serum batches [5]
- Overcrowding and dome formation, which occurs when fluid accumulates beneath the cell layer, creating uneven treatment distribution and inadequate oxygen supply [3]
Monolayer Integrity and Experimental Variability
Q: How can I ensure my Caco-2 monolayers have properly formed before experiments?
A: Caco-2 cells require 21 days post-seeding to establish fully differentiated, stable monolayers [1] [6] [4]. Verify monolayer integrity through:
- Transepithelial Electrical Resistance (TEER): Measure regularly; consistent TEER values indicate tight junction formation [6] [4].
- Paracellular Marker Flux: Use Lucifer Yellow (LY) to assess tight junction integrity; low LY permeability indicates proper monolayer formation [6].
- Microscopy: Visualize zonula occludens-1 (ZO-1) staining to confirm tight junction distribution [6].
- Quality Control: Include reference compounds in each experiment to monitor system performance over time [7].
Q: My permeability results show high variability between experiments. How can I improve consistency?
A: Caco-2 cells exhibit inherent variability due to their heterogeneous nature [4]. Improve consistency by:
- Standardizing Passage Number: Limit continuous cultures to three months and monitor for phenotypic drift [3].
- Controlling Culture Conditions: Use consistent serum batches, media composition, and seeding densities [1] [8].
- Validating Monolayer Integrity: Only use monolayers meeting predefined TEER and LY flux criteria [6] [8].
- Internal Standards: Include calibration compounds with known permeability in each experiment to normalize data [7] [8].
Protocol Optimization and Throughput
Q: The 21-day differentiation period severely limits my throughput. Are there accelerated protocols?
A: Yes, several accelerated models exist but require validation:
- Coated Filters and Supplemented Media: Collagen-coated filters with optimized serum-free media can reduce differentiation to 3-7 days [1] [9].
- Puromycin Treatment: Can enable usable monolayers in 6 days with comparable efflux ratios to traditional models [1].
- Three-Dimensional ECM Substrates: Combined with differentiation media (e.g., butyric acid serum-free DMEM), can produce functional monolayers in 7 days [1].
- Critical Consideration: Accelerated models may have altered transporter expression and barrier integrity; validate against traditional models for your specific applications [1] [6].
Q: Can I re-use Caco-2 monolayers for multiple permeability assays?
A: Yes, with proper recovery periods. Research shows:
- Monolayers maintain morphofunctional properties from day 21 to day 30 [6].
- Permeability assay manipulation causes a small TEER decrease that recovers fully after 2-day incubation with culture media [6].
- Monolayers can be used for assays on days 22, 25, and 28, tripling throughput while maintaining integrity [6].
- Always verify monolayer integrity between uses via TEER and marker flux measurements [6].
Caco-2 Validation for Regulatory Applications
For formal Biopharmaceutics Classification System (BCS) classification and regulatory submissions, Caco-2 validation must demonstrate correlation between apparent permeability coefficient (Papp) and human intestinal absorption (fa) using model drugs spanning permeability ranges [8]. The FDA and EMA require testing at least five model drugs from each permeability category [8].
Table 1: Model Drugs for Caco-2 Validation According to Regulatory Standards
| Permeability Group | Human Absorption (fa) | Example Drugs | Target Papp (×10⁻⁶ cm/s) |
|---|---|---|---|
| High Permeability | ≥85% | Antipyrine, Caffeine, Ketoprofen, Metoprolol | >10 |
| Moderate Permeability | 50-84% | Chlorpheniramine, Terbutaline, Atenolol, Ranitidine | 1-10 |
| Low Permeability | <50% | Famotidine, Nadolol, Acyclovir, Mannitol | <1 |
| Zero Permeability | 0% | FITC-Dextran, Polyethylene glycol 400 | - |
Table 2: Troubleshooting Guide for Common Caco-2 Experimental Issues
| Problem | Potential Causes | Solutions |
|---|---|---|
| Poor Cell Adhesion | Low serum concentration, alkaline medium, insufficient digestion | Maintain 20% FBS, check medium pH, ensure proper trypsinization |
| Slow Growth | Mycoplasma contamination, inadequate NEAA, high passage number | Test for contamination, supplement with NEAA, limit passages |
| High Variability in Permeability | Inconsistent passage practice, varying differentiation levels, genetic drift | Standardize culture protocols, use consistent passage numbers, include internal standards |
| Unstable TEER | Incomplete differentiation, contaminated media, damaged monolayers | Extend differentiation time, use fresh media, handle inserts carefully |
| Unexpected Efflux Ratios | Altered transporter expression, inhibitor contaminants, passage effects | Characterize transporter expression, verify compound purity, control passage number |
Advanced Applications in Permeability Optimization
Integration with Machine Learning and Molecular Pair Analysis
Recent advancements combine Caco-2 data with computational approaches to enhance prediction accuracy and guide molecular optimization:
- Machine Learning Algorithms: XGBoost and other boosting algorithms trained on large Caco-2 datasets can reliably predict permeability, aiding early drug discovery [10].
- Matched Molecular Pair Analysis (MMPA): Identifies chemical transformations that improve permeability while maintaining potency, providing medicinal chemistry guidance [10].
- Transferability Assessment: Models trained on public data show reasonable predictive efficacy when applied to pharmaceutical industry datasets, facilitating knowledge transfer [10].
Machine Learning and Molecular Pair Analysis Workflow for Caco-2 Permeability Optimization
Beyond Traditional Monocultures: Advanced Model Systems
To address Caco-2 limitations, researchers developed enhanced models that better recapitulate intestinal physiology:
- Co-culture Models: Incorporating mucin-producing HT29-MTX cells creates a mucus layer, improving physiological relevance for compounds affected by mucusal interaction [9].
- Gut/Liver Microphysiological Systems (MPS): Fluidically linking Caco-2 models with hepatocyte systems enables first-pass metabolism assessment, providing more accurate bioavailability predictions [2].
- Stem Cell-Derived Models: Primary human intestinal epithelial cells from stem cells (e.g., RepliGut) offer more physiologically relevant transporter and metabolic enzyme expression profiles [2].
Table 3: Research Reagent Solutions for Caco-2 Permeability Studies
| Reagent/Category | Function/Application | Examples/Specifications |
|---|---|---|
| Culture Media | Supports cell growth and differentiation | MEM or DMEM with 4.5 g/L glucose, 20% FBS, 1% NEAA, 1% Pen/Strep [5] [4] |
| Filter Inserts | Platform for polarized cell growth | Polycarbonate membrane, 0.4 μm pore size, 1.12 cm² surface area [6] [4] |
| Coating Reagents | Enhances cell adhesion and differentiation | Collagen Type I (1/100 dilution) [4] |
| Permeability Markers | Monolayer integrity assessment | Lucifer Yellow (paracellular), Propranolol (transcellular), FITC-Dextran (zero permeability) [6] [8] |
| TEER Equipment | Barrier integrity measurement | Epithelial voltohmmeter, chopstick electrodes [6] [4] |
Standard Caco-2 Monolayer Preparation and Permeability Assay Workflow
While the Caco-2 model remains the gold standard for predicting intestinal permeability, researchers must understand its limitations and implement appropriate troubleshooting strategies. The model's tendency toward tighter tight junctions than human small intestine, variable transporter expression, and limited metabolic capability necessitate careful experimental design and interpretation [1] [2]. Nevertheless, through standardized protocols, proper validation, and integration with emerging technologies like machine learning and microphysiological systems, the Caco-2 model continues to provide invaluable insights for drug development and molecular optimization research.
Future directions point toward more physiologically complex models while maintaining the reproducibility and ease of use that established Caco-2 as a pharmaceutical industry standard. By understanding both the capabilities and limitations of this workhorse model, researchers can effectively troubleshoot experimental challenges and generate reliable, predictive permeability data to advance drug development programs.
Understanding Matched Molecular Pair Analysis (MMPA) and Its Power in Drug Design
Frequently Asked Questions (FAQs)
Q1: What is a Matched Molecular Pair, and why is it fundamental to MMPA? A Matched Molecular Pair (MMP) is defined as two compounds that are identical except for a single, well-defined structural transformation at one site [11]. This concept is the cornerstone of MMPA, as it allows scientists to directly correlate a specific chemical change with a resulting change in a biological or physicochemical property, such as Caco-2 permeability [12]. By isolating this single variable, researchers can build causal relationships that guide molecular optimization.
Q2: Our experimental dataset is relatively small. Can we still perform meaningful MMPA? Yes, you can. A powerful approach for small datasets is the MMPA-by-QSAR paradigm [11]. This method involves:
- Building a high-quality QSAR model using your small experimental dataset.
- Using this model to predict properties for a larger, virtual library of compounds.
- Performing MMPA on this mixed dataset (experimental and credible predicted data). This workflow allows you to explore infrequent chemical transformations and generate more design ideas that would not be possible with your small experimental dataset alone [11].
Q3: When I analyze a common transformation, the average effect is near zero. How should I interpret this? This is a common observation, particularly for biological activity endpoints [13]. An average change near zero often indicates that the effect of the transformation is highly context-dependent. The overall distribution may be symmetrical, but within a specific molecular scaffold or protein binding site, the effect could be consistently positive or negative. You should:
- Investigate Context: Analyze if the transformation's effect correlates with specific molecular descriptors or structural environments [13].
- Check Statistical Significance: Ensure you have a sufficient number of pairs and apply statistical tests. Do not rely on the mean value alone if the result is not statistically significant [14] [15].
Q4: How can we ensure that the design rules from public MMPA are applicable to our specific project? The applicability of public data is a key challenge. To improve reliability, you should:
- Seek Statistical Significance: Prioritize transformation rules derived from a large number of pairs and that show statistically significant effects [14] [15].
- Evaluate Contextual Similarity: If possible, compare the structural context (e.g., the atom environment where the transformation occurs) of your project molecules with those in the public database [13]. Rules derived from a similar context are more likely to be transferable.
- Internal Validation: Use your in-house data to validate the most promising public rules before full-scale implementation [16].
Q5: What are the critical statistical considerations for robust MMPA? Ignoring statistics is a major pitfall. Key considerations include:
- Experimental Uncertainty: Account for the noise in your experimental data. Small measured changes may not be statistically significant [14].
- Number of Pairs: The confidence in a derived "rule" increases with the number of matched pairs that support it. Be cautious of rules based on only a few observations [14] [15].
- Y-randomization: This test helps validate your models by ensuring that the observed structure-activity relationships are not due to chance correlations [16].
Troubleshooting Common Experimental Issues
Problem: Inconclusive or noisy results from MMPA.
- Potential Cause 1: Poor Data Quality.
- Solution: Implement a rigorous molecular standardization protocol before analysis. This includes salt removal, tautomer standardization, and neutralization to ensure all compounds are in a consistent chemical state [11].
- Potential Cause 2: Insufficient Data.
- Solution: Apply the MMPA-by-QSAR approach to augment your dataset with credible predicted property values, thereby expanding the chemical space for transformation analysis [11].
Problem: A transformation that worked in one project fails in another.
- Potential Cause: Context Dependency.
- Solution: This highlights that the effect of a transformation is not universal. Use tools that capture the atomic environment around the transformation site. When designing new compounds, prioritize transformations that have a proven track record in a context as similar as possible to your current chemical series [13].
Problem: Too many transformation suggestions to process manually.
- Potential Cause: Unfiltered MMPA Output.
- Solution: Implement a filtering and ranking system. Prioritize suggestions based on:
- Statistical significance of the property change.
- Magnitude of the desired improvement.
- Synthetic feasibility for your team.
- Multi-parameter optimization potential (e.g., does it improve permeability without worsening solubility?) [12].
- Solution: Implement a filtering and ranking system. Prioritize suggestions based on:
MMPA in Action: Optimizing Caco-2 Permeability
The Caco-2 cell assay is a gold standard for predicting intestinal permeability but is time-consuming and costly [16]. MMPA integrates seamlessly into this workflow by providing data-driven hypotheses to improve permeability early in the discovery process.
Workflow: Applying MMPA for Caco-2 Permeability Optimization
The following diagram illustrates how MMPA is integrated into the drug discovery workflow to optimize Caco-2 permeability.
Key Statistical Concepts for Robust Analysis
When evaluating potential transformations, it is crucial to assess their statistical reliability. The following table outlines key metrics and considerations.
| Concept | Description | Importance for MMPA |
|---|---|---|
| Experimental Uncertainty | The inherent noise or error in the experimental measurement of the property (e.g., Caco-2 Papp value). | A measured change must be significantly larger than the experimental uncertainty to be considered real [14]. |
| Statistical Significance (p-value) | The probability that the observed effect is due to random chance. | A small p-value (e.g., < 0.05) increases confidence that the transformation has a genuine, reproducible effect [14] [15]. |
| Number of Pairs (N) | The count of unique matched pairs that support a specific transformation rule. | Rules based on a larger number of observations (high N) are more robust and reliable than those from a few pairs [14]. |
| Applicability Domain | The chemical space defined by the data used to build a model or rule. | Predictions are more reliable for new compounds that fall within the applicability domain of the original MMPA [16]. |
Research Reagent Solutions: The Computational Toolkit
Successful implementation of MMPA relies on several software tools and resources. The table below lists essential components of the MMPA toolkit.
| Tool / Resource | Function | Role in MMPA |
|---|---|---|
| KNIME | An open-source platform for data analytics and integration. | Provides a visual interface for building semi-automated MMPA workflows, including data preparation, QSAR modeling, and MMP calculation [11]. |
| RDKit | An open-source toolkit for cheminformatics. | Used for molecular standardization, descriptor calculation, and fingerprint generation (e.g., Morgan fingerprints) to represent molecular structures [16] [11]. |
| mmpdb | An open-source matched molecular pair platform. | Systematically fragments molecules to create a database of MMPs and calculates transformation rules from large datasets [17]. |
| QSAR Models | Predictive computational models (e.g., Random Forest, XGBoost). | Used in the MMPA-by-QSAR paradigm to predict properties for virtual compounds, expanding the dataset for analysis [16] [11]. |
| Corporate Database | A centralized collection of in-house chemical structures and assay data. | The most valuable resource; internal data provides project-specific context for generating and validating transformation rules [16] [12]. |
By integrating these FAQs, troubleshooting guides, and structured workflows into your research practice, you can leverage the full power of Matched Molecular Pair Analysis to make smarter, data-driven decisions and accelerate the optimization of Caco-2 permeability in your drug discovery programs.
Key Physicochemical Properties Governing Caco-2 Permeability
Frequently Asked Questions (FAQs)
Q1: What are the key acceptance criteria for verifying Caco-2 monolayer integrity before a permeability assay? To ensure reliable permeability results, the cell monolayer must meet specific quality control standards before beginning an experiment. The acceptance criteria can vary based on the format of the transwell plate used. The following table summarizes the key benchmarks for two common formats [18]:
| Measurement | CacoReady 24w | CacoReady 96w |
|---|---|---|
| Transepithelial Electrical Resistance (TEER) | > 1000 Ω·cm² | > 500 Ω·cm² |
| Lucifer Yellow (LY) Apparent Permeability (Papp) | ≤ 1 x 10⁻⁶ cm/s | ≤ 1 x 10⁻⁶ cm/s |
| LY Paracellular Flux | ≤ 0.5% | ≤ 0.7% |
Q2: How is Caco-2 permeability quantitatively measured and used to predict in vivo absorption? The primary quantitative outcome from a Caco-2 assay is the apparent permeability coefficient (Papp), calculated from the permeation rate and the initial concentration of the compound [18]. The calculated Papp value is then used to predict the compound's likely absorption in the human intestine based on established in vitro/in vivo correlations [18]:
| In vitro Papp values | Predicted In Vivo Absorption |
|---|---|
| Papp ≤ 10⁻⁶ cm/s | Low (0-20%) |
| 10⁻⁶ cm/s < Papp ≤ 10 x 10⁻⁶ cm/s | Medium (20-70%) |
| Papp > 10 x 10⁻⁶ cm/s | High (70-100%) |
Q3: Which reference compounds should I use to validate my Caco-2 permeability assay? Using appropriate reference compounds is crucial for assay validation and for distinguishing between different permeability pathways. It is recommended to use at least a high-permeability and a low-permeability control, and to include compounds for studying active transport mechanisms [18].
| Compound Class | Example Compounds (Suggested Concentration) |
|---|---|
| Low Permeability Control | Atenolol (10 µM) |
| High Permeability Control | Propranolol (10 µM), Metoprolol (10 µM) |
| MDR1 (P-gp) Substrate | Digoxin (10 µM) |
| MDR1 (P-gp) Inhibitor | Verapamil (10 µM) |
| BCRP Substrate | Prazosin (1 µM) |
| BCRP Inhibitor | Ko143 (1 µM) |
Q4: My compound shows a large discrepancy between Caco-2 permeability and its observed oral bioavailability. What could explain this? This is a common challenge, often indicating the involvement of transporters or metabolism not fully captured in a standard Caco-2 model. The Caco-2 cell line expresses various influx and efflux transporters (e.g., P-glycoprotein). A compound that is a substrate for an efflux transporter will show lower apparent permeability in the A-to-B direction, which may not reflect its true passive diffusion potential [18] [19]. Furthermore, standard Caco-2 models lack a mucosal layer and may not fully replicate the metabolic environment of the human intestine [9]. To troubleshoot, conduct a bidirectional assay (A-to-B and B-to-A). A high efflux ratio (B-to-A Papp / A-to-B Papp > 2-3) suggests active efflux is limiting absorption [18].
Q5: How can I improve the throughput of my Caco-2 permeability screening without sacrificing data quality? While the traditional Caco-2 assay is low-throughput due to a 21-day differentiation period, several strategies can enhance efficiency [9] [20]:
- Use Ready-to-Use Plates: Commercially available pre-differentiated Caco-2 monolayers (e.g., CacoReady) can reduce preparation time [18].
- Adopt Real-Time Analyzers: Implement impedance-based systems like the xCELLigence RTCA, which allows for non-invasive, real-time monitoring of monolayer integrity and compound effects without manual TEER measurements [20].
- Leverage In-Silico Models: For early-stage compound prioritization, use validated machine learning models to predict Caco-2 permeability based on chemical structure, reserving lab-based assays for later stages [21] [10] [16].
Troubleshooting Guides
Monolayer Integrity Issues
Problem: TEER values are too low or do not reach the required threshold, indicating a leaky monolayer.
| Possible Cause | Recommended Solution |
|---|---|
| Incorrect cell culture conditions | Ensure cells are between passage 30-50. Change culture medium every 2 days and allow a full 15-21 days for differentiation [18] [20]. |
| Microbial contamination | Implement strict aseptic techniques and regularly test for mycoplasma. |
| Toxic compounds or solvents in assay | Verify that the concentration of solvents like DMSO does not exceed 1% (v/v). Include a vehicle control to assess solvent toxicity. |
High Variability in Replicate Measurements
Problem: Triplicate Papp measurements for the same compound show unacceptably high standard deviation.
| Possible Cause | Recommended Solution |
|---|---|
| Inconsistent monolayer quality | Use a real-time cell analyzer (e.g., xCELLigence) to pre-qualify plates with uniform CI values before the assay, ensuring consistent monolayers across all wells [20]. |
| Inaccurate liquid handling | Use calibrated pipettes and consider automated liquid handling systems to improve precision during sampling and dosing. |
| Compound instability or adhesion | Check the compound's stability in the assay buffer. Use mass spectrometry for concentration analysis to avoid interference from compound degradation [18] [22]. |
Poor Correlation with In Vivo Data
Problem: Compounds with high Caco-2 Papp show poor in vivo absorption, or vice versa.
| Possible Cause | Recommended Solution |
|---|---|
| Overlooking active transport | Perform bidirectional assays to identify efflux. Use specific transporter inhibitors (e.g., Verapamil for P-gp) to confirm transporter involvement [18] [19]. |
| Model lacks physiological relevance | Consider using advanced co-culture models, such as Caco-2/HT29-MTX, which incorporates a mucus layer for a more accurate simulation of the intestinal environment [9]. |
| Aqueous solubility issues | Ensure the compound is fully soluble in the assay buffer at the test concentration. Precipitation can lead to an underestimation of permeability. |
Experimental Protocols & Workflows
Standard Caco-2 Permeability Assay Protocol
This protocol outlines the key steps for performing a permeability assay using ready-to-use differentiated Caco-2 monolayers [18].
Workflow Overview
Detailed Methodology
- Step 1: Monolayer Integrity Check: Upon receiving pre-cultured plates, verify monolayer integrity by measuring TEER. Accept only wells meeting the criteria (e.g., TEER > 1000 Ω·cm² for 24-well plates). Alternatively, use Lucifer Yellow (LY) flux as a functional integrity test [18].
- Step 2: Compound Preparation: Prepare the test and reference compounds in the appropriate assay buffer (e.g., HBSS). A suggested starting concentration for unknown compounds is 10 µM. Ensure the final solvent concentration is ≤1% [18].
- Step 3: Dosing and Incubation: For A-to-B (apical-to-basal) permeability, add the compound solution to the apical donor compartment and fresh buffer to the basal receiver compartment. For B-to-A studies, reverse the compartments. Perform all assays in triplicate. Place the plate in an incubator (37°C, 5% CO₂) on an orbital shaker (to reduce the unstirred water layer) for the assay duration, typically 2 hours [18].
- Step 4: Sample Collection: At the end of the incubation period, collect samples from both the donor and receiver compartments.
- Step 5: Analytical Measurement: Analyze the compound concentration in the collected samples using a sensitive and specific method, such as Liquid Chromatography with Mass Spectrometry (LC-MS/MS) [18].
- Step 6: Data Analysis: Calculate the Apparent Permeability (Papp) using the formula [18]:
Papp (cm/s) = (dQ/dt) / (A × C₀)
Where:
- dQ/dt = Permeation rate (nmol/s)
- A = Membrane surface area (cm²)
- C₀ = Initial donor concentration (nmol/ml)
Real-Time Impedance Monitoring Workflow
For a dynamic, label-free assessment of monolayer integrity and compound effects, an impedance-based assay can be used [20].
Real-Time Monitoring Process
Detailed Methodology
- Step 1: Seed Caco-2 cells on E-Plate: Seed Caco-2 cells directly onto the gold microelectrodes of a specialized E-Plate. The instrument (e.g., xCELLigence RTCA) is placed inside a standard cell culture incubator [20].
- Step 2: Monitor Cell Index (CI) in real-time until plateau: The instrument automatically measures electrical impedance at set intervals and converts it into a dimensionless parameter called the Cell Index (CI). Monitor the CI until it reaches a stable plateau, indicating the formation of a fully confluent and differentiated monolayer. This typically takes 18-21 days and serves as a continuous, non-invasive replacement for TEER [20].
- Step 3: Treat with compound: Once the CI plateau is established, treat the cells with the test compound.
- Step 4: Monitor CI changes to assess impact: Continue monitoring the CI in real-time. A sharp, rapid drop in CI may indicate cytotoxicity, while a slower, gradual decrease could suggest a disruption of tight junctions and barrier integrity [20].
The Scientist's Toolkit: Essential Research Reagents & Materials
The following table lists key materials and solutions used in Caco-2 permeability experiments [18] [20] [22].
| Item Name | Function / Application |
|---|---|
| CacoReady Plates | Pre-differentiated Caco-2 cell monolayers on transwell inserts, ready for experimentation, reducing culture time [18]. |
| Transwell Inserts | Permeable supports with a polyester filter that create apical and basolateral compartments to mimic the intestinal barrier [18]. |
| xCELLigence RTCA S16 System | An instrument for real-time, label-free monitoring of cell proliferation, morphology, and monolayer integrity via impedance [20]. |
| E-Plate 16 | A 16-well plate with integrated gold microelectrodes for use with the xCELLigence system [20]. |
| Hanks' Balanced Salt Solution (HBSS) | A standard physiological buffer used as the transport medium during permeability assays. |
| Lucifer Yellow (LY) | A fluorescent paracellular marker used to validate the integrity of tight junctions in the cell monolayer [18]. |
| Mass Spectrometry (LC-MS/MS) | An analytical technique for the highly sensitive and specific quantification of test compound concentrations in assay samples [18] [22]. |
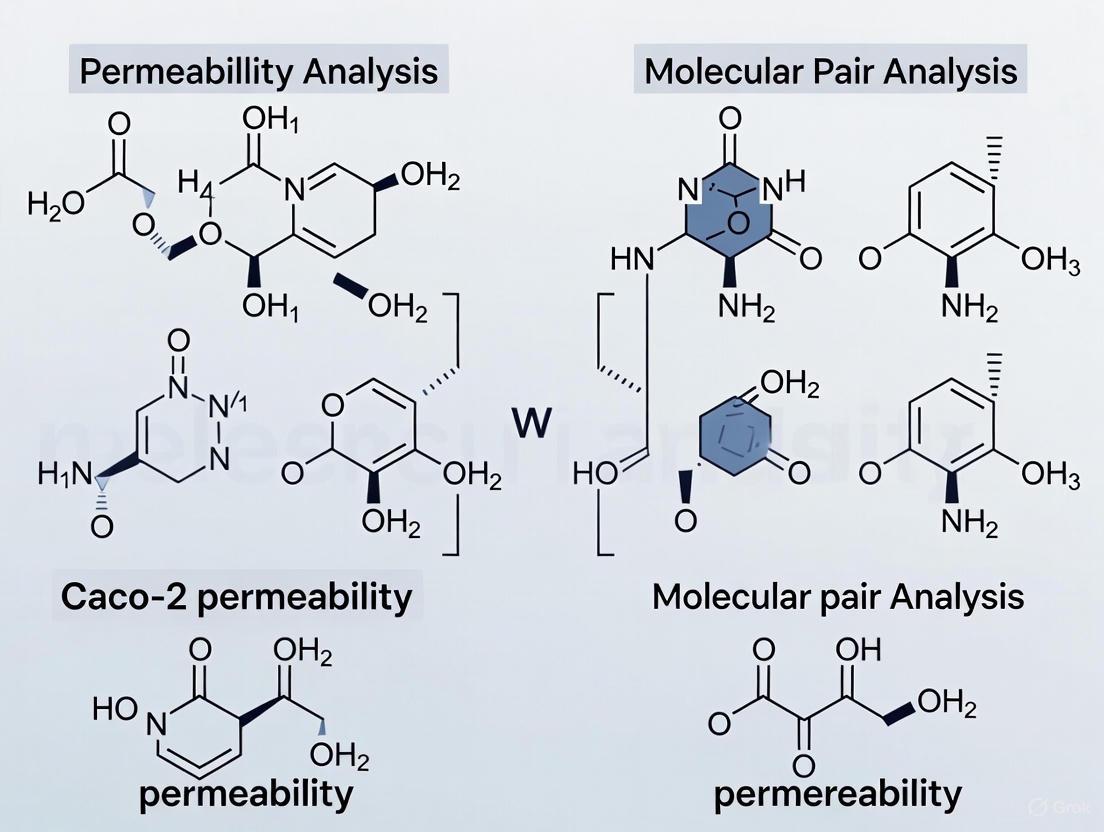
Identifying Common Molecular Transformations that Impact Permeability
FAQs and Troubleshooting Guides
FAQ 1: What is Matched Molecular Pair Analysis (MMPA) and how can it help optimize Caco-2 permeability?
Answer: Matched Molecular Pair Analysis (MMPA) is a computational method that identifies small, specific chemical transformations between pairs of similar compounds and correlates these changes with their experimental property data. In the context of Caco-2 permeability, MMPA extracts chemical transformation rules that provide actionable, quantitative insights for medicinal chemists [16]. By applying these rules, researchers can predict how a specific structural change—such as adding a methyl group or replacing an atom—is likely to increase or decrease a compound's intestinal permeability, thus guiding the rational design of compounds with improved oral absorption [16].
FAQ 2: Why is my computational model for Caco-2 permeability performing poorly on new compound series?
Answer: Poor model generalization, especially for new chemical series (e.g., extended or beyond Rule of 5 space), is a common challenge. This can occur for several reasons [23]:
- Training Data Bias: Global models trained on public data may not perform well on proprietary compound series that occupy a different region of chemical space [16] [23].
- Insufficient Local Context: A single global model might miss subtle, structure-specific relationships that are crucial for a particular project's chemical scaffold [23].
- Experimental Variability: The inherent noise and variability in Caco-2 assay data can hinder the development of highly accurate predictive models [24].
Troubleshooting Guide:
- Interrogate the Applicability Domain: Check if your new compounds fall within the chemical space of the model's training set. If they are outside this domain, predictions will be unreliable [16].
- Explore Local Modeling: Investigate creating a local model for your project. This involves training a model using only compounds that are structurally similar to your series of interest, which can sometimes capture local structure-property relationships more effectively than a global model [23].
- Validate on Internal Data: Always validate the performance of any model, whether public or internally developed, on a held-out test set from your own organization's data to ensure its predictive power is fit for your purpose [16] [23].
FAQ 3: How can I ensure my experimental Caco-2 permeability data is suitable for computational modeling?
Answer: The accuracy and consistency of experimental data are paramount for building reliable computational models [24]. Common pitfalls in experimental data that can derail modeling efforts include:
- Aqueous Boundary Layer (ABL) Dominance: For many compounds, the measured apparent permeability (Papp) is dominated by diffusion through the unstirred water layers rather than by transport across the cell membrane itself. This masks the true intrinsic membrane permeability (P0) [25].
- Low Recovery Rates: Compounds with recovery rates outside the acceptable range (e.g., less than 50% or more than 200%) indicate issues like compound instability or binding to apparatus, making the Papp value unreliable [23].
- Impact of Active Transport: Involvement of efflux transporters (e.g., P-gp) or influx transporters can significantly alter Papp values, complicating the interpretation of passive diffusion mechanisms [24].
Troubleshooting Guide:
- Extract Intrinsic Permeability: Where possible, use specialized kinetic models to extract the intrinsic membrane permeability (P0) from your Papp data, as P0 is a more consistent parameter that eliminates setup-specific factors [25].
- Apply Strict Filters: Before modeling, curate your dataset by excluding compounds with low recovery, significant efflux ratios, or those whose transport is likely limited by the aqueous boundary layer [25] [23].
- Standardize Protocols: Use consistent experimental conditions (e.g., pH, BSA concentration, passage number) across all assays to minimize variability [23] [18].
Key Experimental Data and Molecular Transformations
The following table summarizes quantitative data on the impact of specific molecular transformations on Caco-2 permeability, derived from matched molecular pair analysis and machine learning studies. These rules can serve as a guide for medicinal chemists during compound optimization.
Table 1: Common Molecular Transformations and Their Impact on Caco-2 Permeability
| Molecular Transformation | Typical Impact on Caco-2 Permeability | Notes / Mechanistic Insight |
|---|---|---|
| Introduction of a methyl group (e.g., on an aromatic ring) | Increase [16] | Can reduce polar surface area, improve lipophilicity, or lock a flexible molecule into a more favorable conformation for membrane passage [24]. |
| Cyclization (forming a ring from a chain) | Increase [16] | Often reduces the number of rotatable bonds, which is favorably correlated with improved permeability [24]. |
| Replacement of a carboxylic acid (-COOH) with a bioisostere (e.g., tetrazole, acyl sulfonamide) | Increase [16] | Reduces the number of hydrogen bond donors and the overall charge at physiological pH, facilitating passive transcellular diffusion [24]. |
| Introduction of a hydrogen bond donor (e.g., -OH, -NH₂) | Decrease [16] | Increases the energy penalty for desolvation as the compound partitions into and moves through the lipophilic cell membrane [24]. |
| Increase in molecular weight / size | Decrease (especially beyond 500 Da) [24] | Can hinder transcellular diffusion and is a key parameter in Lipinski's Rule of Five for predicting oral absorption [24]. |
Detailed Experimental Protocol: MMPA for Permeability Optimization
This protocol outlines the key steps for performing a Matched Molecular Pair Analysis to identify permeability-governing transformations, based on methodologies from recent literature [16] [23].
Objective: To systematically identify and quantify the effect of small chemical transformations on Caco-2 permeability within a congeneric compound dataset.
Required Inputs: A curated dataset of chemical structures (e.g., as SMILES strings) and their corresponding experimental Caco-2 Papp values (preferably log-scaled).
Methodology:
Data Curation and Preparation
- Standardize Structures: Use a tool like RDKit to standardize all molecular structures, ensuring consistent tautomer and neutral form representation [23] [26].
- Consolidate Measurements: For compounds with multiple Papp values, calculate the mean value, retaining only those with low standard deviation (e.g., ≤ 0.3) to ensure data reliability [16].
- Apply Data Filters: Exclude compounds with recovery issues (<50% or >200%) or those known to be strong substrates for active transporters unless that is the specific focus of your study [23].
Identification of Matched Molecular Pairs
- Define the Core and R-groups: For your compound series, identify a common molecular core and define the variable R-group attachment points.
- Generate Pairs: Algorithmically identify all pairs of compounds that differ only by a single, well-defined chemical transformation at one of the R-group positions [16].
Calculation of Permeability Change (ΔPapp)
- For each validated molecular pair (A, B), calculate the difference in their experimental permeability values:
- ΔPapp = logPapp(B) - logPapp(A)
- Where the transformation is defined as A → B [16].
- For each validated molecular pair (A, B), calculate the difference in their experimental permeability values:
Statistical Analysis and Rule Extraction
- Aggregate Transformations: Group all pairs that share the identical chemical transformation.
- Calculate Statistics: For each unique transformation, calculate the mean ΔPapp, its standard deviation, and the frequency of occurrence.
- Define Significant Rules: Establish a significance threshold (e.g., a mean |ΔPapp| > 0.3 and a frequency > 5) to filter out noisy or unreliable transformations. The resulting list forms your set of predictive transformation rules [16].
Experimental and Computational Workflow
The diagram below illustrates the integrated workflow for optimizing Caco-2 permeability, combining experimental assays and computational modeling as described in the FAQs and protocols.
Caco-2 Permeability Optimization Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials and Tools for Caco-2 Permeability Research
| Item | Function / Description | Example Use Case |
|---|---|---|
| Caco-2 Cell Line | A human colon adenocarcinoma cell line that, upon differentiation, forms polarized monolayers with functional and structural characteristics of enterocytes [9] [18]. | The "gold standard" in vitro model for predicting human intestinal drug permeability and absorption [9] [3]. |
| Transwell Inserts | Permeable supports with a porous membrane that are placed in multi-well plates. They provide independent access to apical and basolateral compartments, allowing for the creation and study of cell barriers [18]. | Used as the physical scaffold for culturing Caco-2 cells into confluent, differentiated monolayers for permeability assays [18]. |
| TEER Measurement System | Measures Transepithelial Electrical Resistance, a quantitative technique to assess the integrity and tight junction formation of cell monolayers [3] [18]. | Used to validate the quality and confluency of the Caco-2 monolayer before and after permeability experiments. A high TEER value indicates a tight, intact barrier [18]. |
| Reference Compounds (e.g., Propranolol, Atenolol, Digoxin) | Compounds with well-established high, low, or transporter-mediated permeability profiles. They serve as positive and negative controls for the assay [18]. | Used to validate the performance of each assay batch. For example, Propranolol (high permeability) and Atenolol (low permeability) confirm the system's ability to discriminate permeability classes [18]. |
| RDKit | An open-source cheminformatics toolkit that provides functionality for manipulating chemical structures, calculating molecular descriptors, and generating fingerprints [23] [26]. | Used for standardizing molecular structures, calculating descriptors for QSPR model building, and performing molecular pair analysis [23]. |
| LightGBM / XGBoost | Powerful, scalable machine learning algorithms based on gradient boosting frameworks. They are highly effective for building predictive models on structured/tabular data [16] [23] [26]. | Often the top-performing algorithms for building global QSPR models to predict Caco-2 permeability from molecular structures and descriptors [23] [26]. |
From Theory to Practice: Applying Molecular Pair Analysis to Optimize Caco-2 Data
Frequently Asked Questions
FAQ: What is the primary purpose of using Matched Molecular Pair Analysis (MMPA) in Caco-2 permeability studies? MMPA is used to systematically identify specific, small chemical transformations that lead to a predictable change in Caco-2 permeability. This provides data-driven insights for medicinal chemists to rationally optimize a molecule's intestinal absorption potential by suggesting precise structural modifications [16].
FAQ: My machine learning model for Caco-2 permeability performs well on public data but poorly on our in-house dataset. What could be wrong? This is a common issue related to model transferability. It often arises from differences in the structural diversity of compounds or variations in experimental protocols between public and private datasets. To improve performance, consider retraining the model on a combined dataset or using transfer learning techniques. The XGBoost algorithm has shown a good degree of predictive efficacy when applied to industrial data in validation studies [16].
FAQ: How can I assess the reliability of a Caco-2 permeability prediction for a new compound? Implement an Applicability Domain (AD) analysis. This assessment determines whether a new compound falls within the chemical space of the compounds used to train the model. Predictions for molecules outside the model's applicability domain should be treated with caution, as the model may not be reliable for those structures [16].
Troubleshooting Guide: Common Issues in Caco-2 Permeability Prediction Workflows
| Problem Area | Specific Issue | Potential Root Cause | Corrective & Preventive Actions |
|---|---|---|---|
| Data Quality | High variability in permeability measurements for duplicates. | Inconsistent experimental conditions or compound purity. | Apply data curation: exclude duplicates with standard deviation > 0.3 log units [16]. |
| Model Performance | Poor performance on new, proprietary compounds. | Dataset shift between public training and private validation sets. | Use algorithms like XGBoost known for better transferability and perform applicability domain analysis [16]. |
| Chemical Insights | Difficulty translating model results into design rules. | Lack of interpretability in complex machine learning models. | Perform Matched Molecular Pair Analysis (MMPA) to extract specific chemical transformation rules [16]. |
| Model Robustness | Model gives high predictions for impossible structures. | Model learned chance correlations rather than true structure-property relationships. | Conduct a Y-randomization test to validate the model is learning real patterns [16]. |
Experimental Protocol: Building a Model for Chemical Transformation Rules
This section details the methodology for developing a machine learning model to predict Caco-2 permeability and subsequently extract chemical transformation rules via MMPA [16].
Step 1: Data Collection and Curation
- Data Compilation: Collect experimental Caco-2 permeability (Papp) values from multiple public datasets. An initial dataset of 7,861 compounds was used in the referenced study.
- Unit Conversion and Standardization: Convert all permeability measurements to consistent units (e.g., cm/s × 10–6) and apply a logarithmic (base 10) transformation for modeling.
- Data Cleaning:
- Remove entries with missing permeability values.
- For duplicate entries, calculate the mean and standard deviation. Retain only those duplicates with a standard deviation ≤ 0.3 log units, using the mean value for modeling.
- Use a tool like the RDKit MolStandardize module to perform molecular standardization, ensuring consistent tautomer and neutral forms.
- Dataset Splitting: Randomly split the final curated dataset (e.g., 5,654 compounds) into training, validation, and test sets, typically in an 8:1:1 ratio. To ensure robustness, repeat this splitting process multiple times (e.g., 10 times) with different random seeds.
Step 2: Molecular Representation Choose one or more of the following methods to convert chemical structures into a machine-readable format:
- Morgan Fingerprints: Use a radius of 2 and 1024 bits (RDKit implementation).
- RDKit 2D Descriptors: A set of standardized numerical descriptors representing molecular properties.
- Molecular Graphs: Represent molecules as graphs where atoms are nodes and bonds are edges. This is used for graph-based neural networks like DMPNN.
Step 3: Model Construction and Training Train multiple machine learning algorithms to predict the log-transformed Papp values. The study found that XGBoost generally provided superior predictions [16].
- Algorithms to Compare: XGBoost, Random Forest (RF), Support Vector Machine (SVM), and deep learning models like DMPNN.
- Validation: Use the validation set for hyperparameter tuning and the test set for final evaluation. Key performance metrics include R² (coefficient of determination) and RMSE (Root Mean Square Error).
Step 4: Model Validation with Y-Randomization and Applicability Domain
- Y-Randomization Test: Shuffle the permeability values (the Y-vector) across different compounds and attempt to retrain the model. A robust model should fail to learn, showing low performance. This confirms the model is learning true structure-property relationships and not chance correlations.
- Applicability Domain (AD) Analysis: Define the chemical space of the training set. Use a method like the leverage approach to determine if a new compound's prediction is reliable based on its similarity to the training data.
Step 5: Extracting Rules with Matched Molecular Pair Analysis (MMPA)
- Identify Pairs: Systematically identify pairs of compounds within your dataset that differ only by a single, well-defined chemical transformation at a specific site (e.g., -H → -F, -CH₃ → -OCH₃).
- Calculate Permeability Change: For each matched pair, calculate the average change in Caco-2 permeability (ΔPapp) caused by that transformation.
- Derive Rules: Statistically analyze the transformations to identify those that consistently lead to a significant increase or decrease in permeability. These become your data-driven chemical transformation rules for optimization.
Research Reagent Solutions
The following table lists key computational tools and data used in the workflow [16].
| Item Name | Function / Application |
|---|---|
| RDKit | An open-source cheminformatics toolkit used for molecular standardization, fingerprint generation (Morgan), and descriptor calculation (RDKit 2D). |
| XGBoost | A machine learning algorithm based on gradient boosting, identified as providing high predictive accuracy for Caco-2 permeability in the referenced study. |
| Curated Public Caco-2 Dataset | A high-quality, consolidated dataset of Caco-2 permeability measurements for model training and validation. |
| ChemProp | An open-source package used to implement Directed Message Passing Neural Networks (DMPNN) for molecular property prediction. |
| Matched Molecular Pair (MMP) Algorithm | A computational method to fragment and index molecules in a dataset to systematically find all pairs that differ by a single structural change. |
Workflow Visualization
Molecular Pair Analysis Logic
Integrating MMPA with High-Throughput Caco-2 Assay Workflows
In modern drug discovery, the integration of computational and experimental methods is paramount for enhancing efficiency and predictive power. This guide focuses on the practical integration of Matched Molecular Pair Analysis (MMPA) with high-throughput Caco-2 permeability assays. Caco-2 cells, derived from human colon adenocarcinoma, form a monolayer that mimics the human intestinal epithelium, making them a "gold standard" for predicting intestinal absorption and oral bioavailability of drug candidates [27] [8]. However, the traditional Caco-2 assay is time-consuming, requiring extended culturing periods of 7–21 days for full differentiation, which poses challenges for high-throughput screening [28] [8]. MMPA, a computational technique that identifies systematic chemical transformations and their effects on properties, can optimize this process by predicting how specific structural changes will impact Caco-2 permeability before synthesis and testing [28]. This integration allows researchers to prioritize the most promising compounds, guide rational design, and ultimately accelerate the lead optimization process. The following sections provide a technical support framework, including key reagents, troubleshooting guides, and FAQs, to help researchers successfully implement this synergistic workflow.
Key Research Reagent Solutions
The table below lists essential reagents and materials required for establishing and validating the Caco-2 permeability assay, which forms the experimental core of the integrated workflow.
Table 1: Essential Reagents and Materials for Caco-2 Permeability Assays
| Item | Function/Description | Example Usage & Notes |
|---|---|---|
| Caco-2 Cell Line | Human colon adenocarcinoma cell line that differentiates into enterocyte-like cells, forming a polarized monolayer with tight junctions and microvilli [8]. | The foundation of the in vitro model. Use consistent passage numbers and source to minimize variability. |
| Transwell Inserts | Permeable supports with a polyester filter, providing independent access to apical and basolateral compartments to mimic the intestinal lumen and blood circulation [18]. | Available in 24-well and 96-well formats. The surface area is a critical factor in Papp calculations [18]. |
| Validation Compounds | A set of model drugs with known permeability and human absorption values, required for calibrating and validating the Caco-2 model [8]. | Includes high (e.g., Propranolol, Metoprolol), moderate, and low permeability (e.g., Atenolol) compounds, as well as efflux substrates (e.g., Digoxin) [8] [18]. |
| Transporter Inhibitors | Pharmacological agents used to identify the involvement of specific efflux transporters like P-glycoprotein (P-gp) or BCRP [18] [29]. | Examples: Verapamil (P-gp inhibitor), Ko143 (BCRP inhibitor). Used in bidirectional assays to confirm efflux mechanisms [18]. |
| Integrity Markers | Compounds like Lucifer Yellow (LY) used to verify the integrity and confluence of the cell monolayer before and during the permeability assay [18] [29]. | A paracellular flux index (LY Papp) of ≤ 1 x 10⁻⁶ cm/s is a typical acceptance criterion for a intact monolayer [18]. |
| Cell Culture Medium | Specialized medium, often DMEM-based, supplemented with serum and other factors, to support cell growth and differentiation over 15-21 days [18]. | Medium changes are typically performed every second day until a confluent, differentiated monolayer is formed [18]. |
Core Experimental Protocols
Standardized High-Throughput Caco-2 Permeability Assay
A robust and reliable Caco-2 assay protocol is the foundation for generating high-quality data that can be effectively paired with MMPA.
1. Cell Culturing and Monolayer Preparation:
- Seeding: Seed Caco-2 cells onto Transwell inserts at a standardized density. For a 96-well format, a typical seeding density is 50,000 cells per well [18] [8].
- Differentiation: Culture the cells for 15-21 days at 37°C, changing the medium every 48 hours. This extended period allows the cells to fully differentiate and form tight junctions [8] [18].
- Standardization: Use consistent culture conditions (passage number, serum batch, media composition) to minimize inter-laboratory and intra-laboratory variability, which is a significant challenge for this assay [8].
2. Monolayer Integrity Validation:
- Transepithelial Electrical Resistance (TEER): Measure TEER values before initiating the permeability assay. Acceptance criteria are typically >500 Ω·cm² for 96-well plates and >1000 Ω·cm² for 24-well plates [18].
- Paracellular Flux Assay: Use an integrity marker like Lucifer Yellow (LY). Accept the monolayer if the apparent permeability (Papp) of LY is ≤ 1.0 × 10⁻⁶ cm/s and the paracellular flux is ≤ 0.7% [18].
3. Permeability Assay Execution:
- Compound Application: Add the test compound to either the apical (for A-B transport) or basolateral (for B-A transport) compartment. A suggested initial concentration for unknown compounds is 10 µM, and it is recommended to run each compound in triplicate in both directions [18] [29].
- Incubation: Incubate the plates for 2 hours at 37°C with gentle agitation [29].
- Sampling: After incubation, collect aliquots from both the donor and receiver compartments.
4. Quantification and Data Analysis:
- Analytical Quantification: Analyze compound concentrations using a sensitive method such as LC-MS/MS [29].
- Papp Calculation: Calculate the apparent permeability coefficient (Papp in cm/s) using the formula: ( Papp = \frac{dQ/dt}{A \times C0} ) where:
- Efflux Ratio Calculation: Calculate the efflux ratio as ( Papp\ (B-A) / Papp\ (A-B) ). A ratio > 2 suggests the compound is a substrate for active efflux transporters [29].
- Permeability Classification: Classify compounds based on their Papp (A-B) values as follows [18]:
- Low permeability: Papp ≤ 1.0 × 10⁻⁶ cm/s
- Medium permeability: 1.0 × 10⁻⁶ cm/s < Papp ≤ 10 × 10⁻⁶ cm/s
- High permeability: Papp > 10 × 10⁻⁶ cm/s
In Silico Workflow for MMPA
The computational MMPA workflow extracts meaningful chemical transformations from high-quality Caco-2 data.
1. Data Curation and Preparation:
- Compile a dataset of Caco-2 Papp values for a large and structurally diverse set of compounds. Publicly available datasets can be combined, but rigorous curation is essential [28].
- Standardize molecular structures (e.g., using RDKit's MolStandardize) to ensure consistent tautomer and neutral forms [28].
- Convert permeability measurements to a consistent unit (e.g., logPapp) and handle replicates by retaining only entries with low standard deviation (e.g., ≤ 0.3) and using their mean value [28].
2. Matched Molecular Pair Identification:
- Use computational tools to systematically identify Matched Molecular Pairs (MMPs)—pairs of compounds that differ only by a single, well-defined structural transformation at a specific site [28].
3. Transformation Analysis and Rule Extraction:
- For each unique molecular transformation (e.g., -H → -F, -CH₃ → -OCH₃), calculate the average change in logPapp (ΔlogPapp) across all instances of that pair in the dataset.
- Statistically significant and consistent ΔlogPapp values form "chemical transformation rules" that can predict the effect of that specific structural change on permeability [28].
The diagram below illustrates the integrated workflow, showing how the experimental and computational cycles inform and enhance each other.
Troubleshooting Guides & FAQs
Frequently Asked Questions (FAQs)
Q1: Our in-house Caco-2 data does not align well with the predictions from an MMPA model built on public data. What could be the cause? This is a common challenge related to data variability and model transferability. Caco-2 permeability measurements can vary significantly between laboratories due to differences in experimental protocols (e.g., culture time, passage number, assay buffer) [30]. A model trained on public data, which aggregates results from various sources, may not directly translate to your specific internal assay conditions. To mitigate this, it is recommended to fine-tune the model using a portion of your high-quality, consistently measured in-house data to calibrate it to your local context [28].
Q2: How can we ensure our Caco-2 assay data is of high enough quality for reliable MMPA? The accuracy of MMPA is entirely dependent on the quality of the input data. To ensure high data quality:
- Implement Rigorous Validation: Use the recommended set of at least 20 model drugs with high, moderate, and low permeability to validate each assay batch. This ensures your system correctly ranks permeability and generates a calibration curve against human absorption data [8].
- Control for Efflux: Always run bidirectional assays (A-B and B-A) for new compounds to identify if active efflux is involved, which can confound simple passive permeability predictions [29].
- Standardize Protocols: Minimize variability by strictly controlling cell culture conditions, passage number, and assay parameters [8]. Compile consistent Papp data by using identical experimental conditions whenever possible [30].
Q3: What is the simplest way to start integrating MMPA if we have a legacy Caco-2 dataset? Begin with a retrospective analysis. Use your existing legacy dataset of compounds and their measured Papp values to identify matched molecular pairs that are already present within your own chemical series. Analyzing the ΔlogPapp for these pairs can reveal insightful structure-permeability relationships specific to your project's chemical space, providing immediate, actionable guidance for future design without requiring new computational infrastructure [28].
Troubleshooting Guide for Common Experimental Issues
Table 2: Troubleshooting Common Caco-2 Assay Problems
| Problem | Potential Causes | Recommended Solutions |
|---|---|---|
| Low TEER / High LY Flux | - Cells not fully differentiated.- Contamination.- Toxic effect of test compound. | - Extend differentiation time to at least 21 days [8].- Check for microbial contamination.- Perform a cytotoxicity assay prior to permeability testing. |
| High Variability in Papp Values | - Inconsistent monolayer integrity.- Variations in cell passage number or culture conditions.- Analytical error in concentration measurement. | - Strictly monitor and enforce TEER/LY acceptance criteria for every well used [18].- Standardize cell culture protocols and use cells within a defined passage range [8].- Use internal standards and validate analytical methods (e.g., LC-MS/MS) [29]. |
| Poor In Vitro-In Vivo Correlation | - Overlooking the role of efflux transporters or metabolism.- Experimental conditions (pH, buffer) not reflecting physiological state. | - Perform bidirectional assays to calculate an efflux ratio and use specific inhibitors (e.g., Verapamil) to confirm P-gp involvement [18] [29].- Consider using fasted-state simulated intestinal fluid (FaSSIF) as the assay buffer. |
| Inconclusive MMPA Results | - Underlying dataset is too small or not diverse enough.- Permeability mechanism varies within the dataset (e.g., passive vs. active transport). | - Augment your dataset with high-quality public data to increase statistical power [28].- Filter your data by transport mechanism (e.g., analyze passive transcellular diffusion compounds separately from known efflux substrates) [30]. |
The following flowchart provides a structured approach to diagnosing and resolving the common issue of poor correlation between Caco-2 data and computational models.
Essential Data Tables for Analysis and Validation
Table 3: Benchmarking Machine Learning Models for Predicting Caco-2 Permeability
This table summarizes the performance of various modeling algorithms, which can underpin the computational component of the integrated workflow. Performance metrics are Root Mean Square Error (RMSE) and Coefficient of Determination (R²) on independent test sets [28] [31].
| Model Type | Test Set RMSE | Test Set R² | Key Characteristics |
|---|---|---|---|
| Multiple Linear Regression (MLR) | 0.47 [31] | 0.63 [31] | Simple, interpretable baseline model. |
| Support Vector Machine (SVM) | 0.39-0.40 [31] | 0.73-0.74 [31] | Effective for non-linear relationships. |
| Random Forest (RF) | 0.39-0.40 [31] | 0.73-0.74 [31] | Robust to outliers and non-linear data. |
| Gradient Boosting Machine (GBM) | 0.39-0.40 [31] | 0.73-0.74 [31] | High performance, often a top contender. |
| XGBoost | Reported as generally better than comparable models [28] | N/A | A leading boosting algorithm known for high predictive accuracy and speed. |
| SVM-RF-GBM Ensemble | 0.38 [31] | 0.76 [31] | Often achieves superior performance by combining multiple models. |
Table 4: Validation Criteria for Caco-2 Monolayer Integrity and Permeability Classification
This table consolidates the key acceptance criteria for a properly functioning Caco-2 assay, which is critical for generating reliable data for MMPA [18].
| Parameter | Measurement Method | Acceptance Criterion (24-well) | Acceptance Criterion (96-well) | Purpose |
|---|---|---|---|---|
| TEER | Voltmeter/Epithelial Voltohmmeter | > 1000 Ω·cm² [18] | > 500 Ω·cm² [18] | Ensures tight junction formation and monolayer integrity. |
| Paracellular Flux (LY Papp) | Apparent Permeability of Lucifer Yellow | ≤ 1.0 × 10⁻⁶ cm/s [18] | ≤ 1.0 × 10⁻⁶ cm/s [18] | Directly measures leakiness of the monolayer. |
| Permeability Classification (Papp A-B) | Calculated from assay data | High: > 10 × 10⁻⁶ cm/sModerate: 1-10 × 10⁻⁶ cm/sLow: ≤ 1 × 10⁻⁶ cm/s [18] | Same as 24-well [18] | Predicts in vivo absorption potential from in vitro data. |
Leveraging Machine Learning and XGBoost for Enhanced Permeability Classification
Frequently Asked Questions (FAQs)
FAQ 1: Why is my XGBoost model performing poorly on Caco-2 permeability data, and how can I improve it?
Poor performance can often be attributed to several common issues. First, ensure your dataset is sufficiently large and chemically diverse; models built on small datasets (e.g., less than 100 compounds) often struggle with generalization and have a narrow application domain [32]. Second, check your molecular descriptors. Using unstable 3D descriptors can introduce noise, whereas robust 2D descriptors like Morgan fingerprints or RDKit 2D descriptors often provide more stable and accurate predictions [32] [16]. Finally, validate that your modeling process adheres to OECD principles, including proper train/test splits, cross-validation, and defining an applicability domain (AD) to ensure robustness and reliability [32].
FAQ 2: How should I handle categorical molecular features in my XGBoost pipeline?
The recommended method is to use XGBoost's built-in support for categorical data. When using a DataFrame (e.g., pandas), simply convert the relevant columns to the category data type. Then, when initializing your XGBoost classifier or regressor, set the parameter enable_categorical=True. It is also crucial to use a supported tree method like hist and to save the model in JSON format to preserve the categorical information [33]. This allows XGBoost to use an optimal partitioning strategy for categorical splits, which is often more efficient than traditional one-hot encoding [33].
FAQ 3: What is the difference between Gain, Cover, and Frequency in XGBoost feature importance, and which should I trust for interpreting my permeability model?
These three metrics offer different perspectives on feature usage [34]:
- Gain: This is the most relevant for understanding a feature's predictive power. It represents the average improvement in model accuracy (the decrease in loss) brought by the feature each time it is used in a split. A higher Gain means the feature is more important for making correct predictions [34].
- Cover: This metric measures the relative number of observations (data points) related to a feature across all trees. It indicates how often the feature is used to make decisions for a large portion of the data [34].
- Frequency: This is simply the percentage of times a feature appears in all the trees across the model [34].
For interpreting your Caco-2 permeability model, Gain is generally the most important metric as it directly quantifies a feature's contribution to prediction accuracy.
FAQ 4: My model trained on public data performs poorly on our in-house corporate compound library. What can I do?
This is a common challenge related to the transferability of models. To improve performance on your proprietary data:
- Analyze the Applicability Domain (AD): Use a distance-based method, potentially weighted by descriptor importance, to determine if your in-house compounds fall outside the chemical space covered by the public training data. Compounds outside the AD may have unreliable predictions [32] [16].
- Data Consolidation and Retraining: If possible, combine the public data with your in-house data (or a representative sample of it) and retrain the model. This helps the model learn the specific characteristics of your corporate library [16].
- Algorithm Selection: Research indicates that boosting algorithms like XGBoost may retain better predictive efficacy when applied to industrial data compared to other methods, making them a robust choice for such scenarios [16].
Troubleshooting Guides
Issue 1: Error When Loading a Saved XGBoost Model for Inference
Problem: You get an error or unexpected behavior when loading a previously saved XGBoost model to make new predictions.
Solution: This is frequently caused by an environment mismatch or an incorrect serialization method [35].
Use the Correct Serialization Format: Always save models trained with categorical data support using XGBoost's native
save_modelmethod and the JSON format [33].Avoid using Python's
picklemodule for these models, as it may not preserve categorical information reliably.Ensure Environment Consistency: The versions of XGBoost and its dependencies should be identical between the training and inference environments. Use a
requirements.txtfile to document the specific versions [35].Verify Categorical Data Encoding for Inference: When making predictions on new data, ensure that categorical columns in the new DataFrame have the same data types (
category) as the training data. Starting from XGBoost 3.1, the Python interface can often perform automatic re-coding for DataFrame inputs, but consistency is key [33].
Issue 2: Low Feature Importance for Physicochemically Meaningful Molecular Descriptors
Problem: A molecular descriptor known from literature to affect permeability (e.g., related to hydrogen bonding) shows low importance in your XGBoost model.
Solution: The definition of "importance" can vary. XGBoost's built-in importance (Gain) measures a feature's contribution to the model's predictive performance on the training data, which can be influenced by feature cardinality and correlation [36].
- Calculate Multiple Importance Metrics: Instead of relying solely on Gain, also calculate Cover and Frequency to get a complete picture of how the feature is being used [34].
- Use SHAP Values for a More Robust Interpretation: SHAP (SHapley Additive exPlanations) values provide a unified measure of feature impact on the model output for individual predictions. A feature can have a high Gain (used for important splits) but a low mean |SHAP| value if its average effect on the output magnitude is small [36]. Using both methods together gives a fuller picture.
- Check for Highly Correlated Features: If two descriptors convey similar information, XGBoost might use only one of them, making the other seem unimportant. Analyze feature correlation and consider grouping them.
Experimental Protocols & Data
Protocol: Building a QSPR Model for Caco-2 Permeability Using XGBoost
This protocol outlines the steps for creating a robust Quantitative Structure-Property Relationship (QSPR) model, as demonstrated in recent literature [32] [16].
1. Data Collection and Curation
- Source: Collect experimental Caco-2 apparent permeability (Papp) values from public databases like ChEMBL and literature compilations.
- Curation:
- Remove entries with missing or unclear Papp values or SMILES codes.
- For duplicate compounds, calculate the mean Papp value if the standard deviation is low (e.g., ≤ 0.3 log units); otherwise, remove them.
- Standardize molecular structures using a tool like RDKit's
MolStandardizeto achieve consistent tautomer and neutral forms. - Convert Papp values to logarithmic scale (e.g., log10(Papp (cm/s × 10–6))) for modeling.
2. Molecular Featurization
- Calculate molecular descriptors or fingerprints. Common choices include:
- Morgan Fingerprints: (Radius 2, 1024 bits) to capture local atomic environments [16].
- RDKit 2D Descriptors: A set of standardized 2D molecular descriptors.
- Perform feature selection to reduce dimensionality. Methods include:
3. Data Splitting
- Use a combination of Principal Component Analysis (PCA) and Self-Organizing Maps (SOM) to ensure chemically diverse splits into training and test sets, rather than simple random splitting [32].
- A typical split ratio is 80:20 or 80:10:10 for training, validation, and test sets, respectively [16].
4. Model Training and Validation
- Training: Train the XGBoost regressor (
XGBRegressor) on the training set. Hyperparameter tuning is critical. - Validation:
- Internal: Use k-fold cross-validation (e.g., k=5) on the training set to assess robustness [32].
- External: Evaluate the final model on the held-out test set.
- Y-Randomization: Test the model on data where the target (Papp) is randomized. A significant drop in performance confirms the model learned real structure-property relationships and not noise [16].
- Metrics: Report R² (coefficient of determination), RMSE (Root Mean Square Error), and MAE (Mean Absolute Error) for both training and test sets.
Experimental Workflow The diagram below visualizes the key stages of the QSPR modeling workflow.
Key Experimental Data and Performance
The following table summarizes the scope and performance of XGBoost models from recent Caco-2 permeability studies, highlighting the importance of data set size and model validation.
Table 1: Performance of XGBoost Models in Caco-2 Permeability Prediction
| Study Description | Data Set Size (Compounds) | Key Descriptors / Features | Validation Method | Reported Performance (Test Set) |
|---|---|---|---|---|
| QSPR Model with Dual-RBF & XGBoost [32] | 1,827 | PaDEL descriptors, selected via MDI and HQPSO | Train/Test split, series of validations | Dual-RBF (Best): R² = 0.77XGBoost: Part of model comparison |
| Comprehensive ML Algorithm Validation [16] | 5,654 (after curation) | Morgan Fingerprints, RDKit 2D descriptors, Molecular Graphs | 80/10/10 split, 10 independent runs, external industrial set | XGBoost: Generally provided better predictions than comparable models (RF, SVM, GBM) on test sets. |
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Computational Tools and Resources for Caco-2 Permeability Modeling
| Item / Resource | Function / Description | Relevance to Caco-2 Permeability Experiments |
|---|---|---|
| RDKit | An open-source cheminformatics toolkit. | Used for molecular standardization, calculation of 2D descriptors, and generation of Morgan fingerprints [16]. Essential for the featurization step. |
| PaDEL-Descriptors | Software to calculate molecular descriptors and fingerprints. | Used to generate a comprehensive set of 1D and 2D descriptors that serve as input features for the QSPR model [32]. |
| ChEMBL Database | A large-scale bioactivity database for drug discovery. | A primary source for obtaining experimental Caco-2 permeability data for model training [32]. |
| XGBoost Library | An optimized gradient boosting library. | The core machine learning algorithm used to build the regression model that predicts permeability from molecular features [32] [16]. |
| SHAP Library | A game theory-based method to explain model outputs. | Critical for interpreting the XGBoost model, identifying which molecular features drive high or low permeability predictions for specific compounds [36]. |
Frequently Asked Questions: Permeability in bRo5 Space
Q1: What are the key property ranges for orally bioavailable compounds in the bRo5 space? Oral drugs in the bRo5 space occupy a narrow range of properties that balance permeability and solubility. Key limits include a Molecular Weight (MW) up to 1000–1100 Da and a lipophilicity (cLogP) up to 10–13 [37]. It is critical to keep the topological polar surface area (TPSA) proportional to the MW; a TPSA/MW ratio of 0.1-0.3 Ų/Da is a typical target for highly permeable compounds [38].
Q2: What is a "molecular chameleon" and why is it important for bRo5 permeability? A molecular chameleon is a flexible molecule that can change its conformation based on its environment [37]. In aqueous, polar environments, it adopts a more open, polar conformation, which is good for solubility. In apolar, membrane-like environments, it folds into a less polar, more compact conformation by forming intramolecular hydrogen bonds (IMHBs) and other interactions, which is essential for permeability [37]. This chameleonic behavior allows bRo5 compounds to achieve cell permeability that can be nearly two orders of magnitude higher than if they remained in a polar conformation [37].
Q3: My Caco-2 assay shows low permeability. What molecular strategies can I use to improve it? For bRo5 compounds, improving permeability often involves optimizing properties to enhance chameleonicity:
- Reduce 3D Polar Surface Area (PSA): Aim for a 3D PSA below 100 Ų in a membrane-like environment. This is a stronger indicator of permeability than the traditional topological PSA (TPSA) [38].
- Manage Lipophilicity: While bRo5 compounds often have high logP, permeability is best when lipophilicity is balanced within a narrow window, especially as MW increases [37].
- Limit Hydrogen Bond Donors (HBDs): Although oral bRo5 drugs can have up to 6 HBDs, it is desirable to limit them to 2-3, particularly if they are from ureas or amides [37].
- Use Matched Molecular Pair (MMP) Analysis: Systematically identify small chemical transformations that improve permeability. For example, introducing specific lipophilic substituents or adding a polar atom like nitrogen can significantly alter permeability and other ADME properties [39].
Q4: My Caco-2 cells are not forming a proper monolayer, or I have many floating cells. What could be wrong? Caco-2 cells have unique growth characteristics that require specific conditions [5]:
- Slow Adhesion and Growth: Cells can take 24-72 hours to adhere after seeding and may require passaging only once a week. Ensure your culture medium is not alkaline (purple-red), as this hinders adhesion [5].
- Culture Medium: Using MEM without non-essential amino acids (NEAA) can reduce growth rate and increase floating cells. A high concentration of Fetal Bovine Serum (FBS), typically 20%, is often required for proper adhesion and growth [5].
- Digestion Difficulties: These cells are tightly connected and hard to dissociate into single cells. Digestion with trypsin typically takes 5-10 minutes, and it's normal for the cells to detach in clusters rather than as single cells [5].
Troubleshooting Guide: Common Experimental Issues
| Problem | Possible Cause | Solution |
|---|---|---|
| Low Caco-2 Permeability (Papp) | High 3D PSA in membrane environment; Insufficient intramolecular H-bonds; Suboptimal lipophilicity window [38] [37]. | Use conformational analysis to design for lower 3D PSA; Introduce structural motifs that stabilize intramolecular H-bonds; Use MMP analysis to fine-tune logP [38] [39]. |
| Poor Aqueous Solubility | Compound remains in a low-polarity, "closed" conformation in water [37]. | Design compounds with a balance of polarity to favor a more "open," hydrated conformation in aqueous environments (chameleonicity) [37]. |
| High Variability in Caco-2 Data | Monolayer integrity is compromised; Cell culture conditions are suboptimal [18]. | Validate monolayer integrity before assay (TEER > 1000 Ω·cm² for 24-well plates; LY Papp ≤ 1 x 10⁻⁶ cm/s); Use standardized, ready-to-use Caco-2 models like CacoReady to ensure consistency [18]. |
| Caco-2 Cells Not Adhering | Alkaline culture medium; Low FBS concentration [5]. | Check medium color (should be orange-red, not purple); Adjust FBS concentration to 20% [5]. |
Experimental Protocols & Data Interpretation
1. Caco-2 Permeability Assay Protocol
- Cell Culture: Seed Caco-2 cells on transwell inserts and culture for 15-21 days to allow formation of a confluent, differentiated monolayer. Change the medium every two days [18].
- Integrity Check: Before the assay, measure Transepithelial Electrical Resistance (TEER). An acceptable TEER value is > 1000 Ω·cm² for 24-well plates. Also, measure the paracellular flux of Lucifer Yellow (LY); the apparent permeability (Papp) for LY should be ≤ 1 x 10⁻⁶ cm/s [18].
- Permeability Assay:
- Add your test compound (e.g., at a suggested 10 µM for unknowns) to the donor compartment (apical for A-B transport, basal for B-A transport).
- Incubate for 2 hours at 37°C.
- Sample from both donor and receiver compartments.
- Analyze compound concentration using a sensitive method like LC-MS/MS [18].
- Data Calculation: Calculate the apparent permeability (Papp) using the formula:
- Papp = (dQ/dt) / (A × C0)
- Where dQ/dt is the transport rate (nmol/s), A is the membrane area (cm²), and C0 is the initial donor concentration (nmol/mL) [18].
2. Interpreting Caco-2 Papp Values for In Vivo Absorption Use the following table to predict absorption based on your in vitro data [18]:
| In vitro Papp Value | Predicted In Vivo Absorption |
|---|---|
| Papp ≤ 1.0 x 10⁻⁶ cm/s | Low (0-20%) |
| 1.0 x 10⁻⁶ cm/s < Papp ≤ 10 x 10⁻⁶ cm/s | Medium (20-70%) |
| Papp > 10 x 10⁻⁶ cm/s | High (70-100%) |
3. Reference Compounds for Caco-2 Assay Validation Always include control compounds in your assay to validate its performance [18]:
| Function | Compound (Example) |
|---|---|
| High Permeability Control | Propranolol |
| Low Permeability Control | Atenolol |
| MDR1 (P-gp) Substrate | Digoxin |
| MDR1 (P-gp) Inhibitor | Verapamil |
| BCRP Substrate | Prazosin |
| BCRP Inhibitor | Ko143 |
The Scientist's Toolkit: Key Reagents & Materials
| Item | Function / Explanation |
|---|---|
| Ready-to-Use Caco-2 Models (e.g., CacoReady) | Pre-seeded, ready-to-assay plates that ensure monolayer consistency and save cell culture time [18]. |
| MEM Culture Medium with 20% FBS and NEAA | Standard growth medium for maintaining healthy, differentiating Caco-2 cells. NEAA (Non-Essential Amino Acids) are crucial for optimal growth [5]. |
| Hank's Balanced Salt Solution (HBSS) | Standard buffer used as the transport medium during the permeability assay. |
| Reference Compounds (Propranolol, Atenolol, etc.) | Critical for validating the correct functioning of the Caco-2 monolayer and the assay itself [18]. |
| Lucifer Yellow (LY) | A fluorescent marker used to measure paracellular flux and confirm the integrity of the tight junctions in the cell monolayer [18]. |
Property Ranges for Oral bRo5 Drugs & Candidates
Analysis of orally absorbed drugs and clinical candidates in the bRo5 space has established the following property ranges [37]:
| Molecular Property | Typical Range for Oral bRo5 |
|---|---|
| Molecular Weight (MW) | Up to 1000 - 1100 Da |
| cLogP | Up to 10 - 13 |
| Hydrogen Bond Donors (HBD) | Up to 6 (2-3 recommended) |
| Hydrogen Bond Acceptors (HBA) | Up to 14 - 15 |
| Topological Polar Surface Area (TPSA) | Up to 230 - 250 Ų |
| Rotatable Bonds (NRotB) | 5 - 20 |
Matched Molecular Pair (MMP) Analysis Workflow
MMP analysis identifies the effect of small, specific structural changes on a property like permeability. The workflow in tools like KNIME is as follows [39]:
- Data Preparation: Gather and standardize chemical structures and associated property data (e.g., pKi, logP).
- Fragmentation & Pair Identification: Systematically fragment molecules at single non-functional group bonds to identify pairs that differ only at a single site.
- Transformation Analysis: Aggregate all data for each unique chemical transformation (e.g., -H → -Ph) and calculate the median change in the property of interest.
- Visualization & Exploration: Interactively explore transformations, filtering by frequency and the magnitude of the property change to identify meaningful design rules.
MMP Analysis Workflow
Molecular Design Logic for bRo5 Permeability
The following diagram illustrates the logical pathway for designing permeable bRo5 molecules, emphasizing the critical role of molecular chameleonicity.
Design Logic for Permeable bRo5 Molecules
Solving Common Caco-2 and MMPA Challenges: A Troubleshooting Guide
Addressing Low Solubility and Recovery in Caco-2 Assays
Troubleshooting Guides
Guide 1: Diagnosing and Resolving Low Compound Recovery
Q: What does low recovery indicate, and why is it a problem? Low recovery, where the total amount of compound recovered at the end of the experiment is significantly less than the initial amount, is a common issue that can lead to ambiguous or misleading data. It primarily indicates non-specific binding of the compound to the assay plasticware or cellular components, but can also result from poor solubility, compound metabolism by the cells, or accumulation within the cell monolayer [40]. Low recovery can cause an underestimation of both permeability and efflux, as the reduced free concentration in solution means less compound is available for uptake or to be detected as effluxed by transporters [40].
Step-by-Step Diagnostic and Resolution Procedure:
Confirm the Issue: Calculate the percentage recovery using the formula below. While acceptance criteria may vary, a very low recovery (e.g., <80%) warrants investigation [41].
% Recovery = (Total compound in donor and receiver at experiment end / Initial compound present) × 100[40]Identify the Root Cause:
- Suspect Non-Specific Binding (NSB): This is the most prevalent cause for lipophilic compounds. Test for adsorption by incubating the compound in the buffer with the empty transwell plate and analyzing concentration over time.
- Check Aqueous Solubility: Ensure the test concentration does not exceed the compound's solubility limit in the assay buffer.
- Investigate Cellular Metabolism or Accumulation: If NSB and solubility are ruled out, the compound may be unstable or metabolized by the Caco-2 cells.
Implement Solutions:
- Add Bovine Serum Albumin (BSA): Incorporate 0.5% to 1% (w/v) BSA into the transport buffer (both donor and receiver compartments) [40] [41]. BSA acts as a scavenger, blocking non-specific binding sites on the plasticware and improving the aqueous solubility of lipophilic compounds by acting as a solubilizing agent [40].
- Use Other Proteins or Serum: In some cases, adding fetal bovine serum (FBS) or other proteins like mucin can be effective, especially for highly challenging compounds [42].
- Optimize Buffers: Use physiologically relevant buffers like FaSSIF (Fasted State Simulated Intestinal Fluid) in the apical compartment to better mimic the intestinal environment and enhance solubility [42].
Guide 2: Overcoming Challenges with Low Solubility Compounds
Q: My compound has poor aqueous solubility. How can I obtain reliable permeability data? Low solubility directly compromises assay reliability by reducing the available free concentration for permeation, leading to underestimation of permeability and potential false-negative efflux results. The goal is to maintain the compound in solution throughout the experiment without damaging the cell monolayer.
Step-by-Step Optimization Protocol:
Pre-experiment Solubility Assessment:
- Prepare the compound in the intended assay buffer (with and without BSA) at the desired test concentration.
- Incubate the solution with gentle agitation at 37°C for the duration of the planned permeability experiment (e.g., 2 hours).
- Centrifuge and analyze the supernatant via LC-MS/MS to confirm the compound remains in solution.
Modify the Assay Buffer:
- Add BSA (1% w/v): As with recovery issues, BSA is a primary tool for solubilizing lipophilic compounds [40] [41].
- Use FaSSIF: Replacing the standard apical buffer (e.g., HBSS) with FaSSIF introduces bile salts and phospholipids, which can dramatically improve the solubility of poorly soluble drugs [42].
Adjust Experimental Conditions:
- Reduce Test Concentration: Lower the compound concentration to a level below its solubility limit, even if it requires more sensitive analytical detection [41].
- Minimize DMSO: Keep the final DMSO concentration as low as possible (typically ≤0.5-1.0%) to avoid monolayer disruption and artificial permeability enhancement [42].
- Consider a Pre-incubation Step: For "beyond Rule of 5" (bRo5) compounds, a pre-incubation step can help the system reach a steady state, improving data quality for very slow-permeating compounds [41] [42].
Frequently Asked Questions (FAQs)
Q: How does low recovery specifically impact the interpretation of efflux ratios? A low recovery can mask a compound's true efflux potential. If a significant portion of the compound binds to the assay plate, the intracellular concentration available for efflux transporters like P-gp or BCRP is reduced. This can result in a lower-than-expected basolateral-to-apical (B-A) flux, causing an efflux ratio that is artificially low (e.g., <2), leading to the false conclusion that the compound is not an efflux substrate [40].
Q: Beyond BSA, what other assay modifications can help with challenging bRo5 compounds like PROTACs? For very complex molecules such as PROTACs (which are typically bRo5), standard Caco-2 assays often fail. A comprehensive optimized protocol, termed an "equilibrated Caco-2 assay," includes several key modifications [41]:
- Pre-incubation: Adding the compound to the donor compartment for 60-90 minutes before the main assay incubation allows the system to approach steady state.
- Prolonged Incubation Time: Extending the incubation beyond the standard 2 hours can help with detection for very slow-permeating compounds.
- Systematic Use of BSA (1%): Included in the buffer to minimize nonspecific binding and improve recovery.
- Enhanced LC-MS/MS Analytics: Optimizing the analytical method for maximum sensitivity is crucial to detect the low concentrations permeating the monolayer.
Q: What are the key acceptance criteria to ensure my Caco-2 monolayer is functioning correctly before testing a challenging compound? Before beginning any permeability experiment, it is critical to validate the integrity and functionality of the Caco-2 cell monolayer. The following table summarizes common acceptance criteria for validated monolayers.
Table 1: Key Acceptance Criteria for Caco-2 Monolayer Integrity
| Measurement | Purpose | Typical Acceptance Criteria | Source |
|---|---|---|---|
| Transepithelial Electrical Resistance (TEER) | Measures tight junction formation and monolayer integrity. | > 500 Ω·cm² (96-well format); > 1000 Ω·cm² (24-well format) | [18] |
| Lucifer Yellow (LY) Papp | Paracellular flux marker to verify tight junction integrity. | ≤ 1.0 × 10⁻⁶ cm/s | [18] [40] |
| LY Paracellular Flux | Alternative measure of paracellular leakage. | ≤ 0.5% - 0.7% | [18] |
| Reference Compound Papp | Validates functionality for passive and active transport. | High-Permeability Marker (e.g., Propranolol): Papp > 10 × 10⁻⁶ cm/s; Low-Permeability Marker (e.g., Atenolol): Papp < 1 × 10⁻⁶ cm/s | [8] [43] |
Experimental Protocols for Critical Experiments
Protocol 1: Optimized Bidirectional Permeability Assay with BSA
This protocol is designed to maximize recovery and data quality for compounds with low solubility or high non-specific binding, framed within a molecular pair analysis study to compare optimized versus standard conditions.
Methodology:
- Cell Culture: Seed Caco-2 cells (e.g., TC7 clone) onto 0.4 µm polyester transwell inserts at a density of 40,000-125,000 cells per well (for 96-well and 24-well formats, respectively). Culture for 7-21 days until fully differentiated, with regular medium changes [41] [42].
- Buffer Preparation: Prepare HBSS buffer, pH 7.4, supplemented with 1% (w/v) Bovine Serum Albumin (BSA) and 10 mM HEPES. This BSA-supplemented buffer is used for both donor and receiver compartments unless specified otherwise [40] [41].
- Monolayer Integrity Check: On the day of the experiment, measure TEER and/or assess the paracellular flux of Lucifer Yellow (80 µM) to ensure monolayer integrity meets acceptance criteria (see Table 1) [18] [40].
- Bidirectional Transport Study:
- A-B Direction: Add test compound (e.g., 1-10 µM) in BSA-supplemented buffer to the apical (donor) compartment. Fill the basolateral (receiver) compartment with BSA-supplemented buffer.
- B-A Direction: Add test compound in BSA-supplemented buffer to the basolateral (donor) compartment. Fill the apical (receiver) compartment with BSA-supplemented buffer.
- Incubate the plate for 2 hours at 37°C with 5% CO₂ and 100% humidity [18] [40].
- Sample Collection and Analysis:
- Take samples from both donor and receiver compartments at the end of the incubation.
- Quench samples with acetonitrile containing an internal standard (e.g., carbutamide).
- Analyze compound concentrations using a sensitive and optimized LC-MS/MS method [41].
- Data Calculation:
- Calculate the apparent permeability (Papp) in both directions.
- Calculate the Efflux Ratio:
ER = Papp(B-A) / Papp(A-B). - Calculate the % Recovery for both directions.
The workflow for this optimized protocol is summarized in the diagram below.
Optimized Caco-2 Assay Workflow
Protocol 2: Pre-incubation Assay for bRo5 Compounds
This protocol is specifically tailored for measuring the permeability of challenging bRo5 compounds (e.g., PROTACs) close to equilibrium, where standard assays fail.
Methodology:
- Cell Culture and Buffer Preparation: Follow steps 1-3 from Protocol 1.
- Pre-incubation Step:
- Add the test compound solution (in BSA-supplemented buffer) to the donor compartments.
- Fill the receiver compartments with BSA-supplemented buffer.
- Incubate for 60-90 minutes at 37°C [41].
- Main Incubation:
- After pre-incubation, remove the solutions from both donor and receiver compartments.
- Rinse the cell monolayer gently with BSA-supplemented buffer.
- Add fresh compound solution to the donor compartments and fresh BSA-supplemented buffer to the receiver compartments.
- Conduct the main incubation for 60 minutes at 37°C [41].
- Sample Collection and Analysis: Proceed with steps 5 and 6 from Protocol 1.
Research Reagent Solutions
The following table details key reagents and materials essential for implementing the optimized Caco-2 assays described in this guide.
Table 2: Essential Reagents for Optimizing Caco-2 Assays
| Reagent/Material | Function | Key Consideration / Benefit |
|---|---|---|
| Bovine Serum Albumin (BSA) | Reduces non-specific binding to plasticware; improves aqueous solubility of lipophilic compounds. | Critical for achieving high recovery and reliable efflux data for BCS Class II/IV and bRo5 compounds [40] [41]. |
| Transwell Plates (0.4 µm pore) | Provides a semi-porous membrane support for cell growth and polarization. | Polyester membranes are commonly used. The 96-well format enables higher throughput [18] [41]. |
| Caco-2 Cells (e.g., TC7 clone) | The in vitro model of the human intestinal epithelium. | Using a consistent clone and passage number improves inter-assay reproducibility [42]. |
| Lucifer Yellow | A fluorescent paracellular marker used to validate monolayer integrity. | Acceptance threshold: Papp (LY) ≤ 1.0 × 10⁻⁶ cm/s [18] [40]. |
| Reference Compounds (Atenolol, Propranolol) | Low and high permeability standards for assay validation and compound ranking. | Ensure consistent rank-order relationship for BCS classification [18] [8] [43]. |
| Efflux Transporter Inhibitors (e.g., Verapamil, Ko143) | Chemical inhibitors (for P-gp and BCRP, respectively) to confirm transporter involvement. | Used in follow-up studies to mechanistically understand efflux signals [18] [40]. |
| FaSSIF (Fasted State Simulated Intestinal Fluid) | Apical buffer simulating intestinal fluid to enhance compound solubility. | Particularly useful for compounds with poor solubility in standard HBSS buffer [42]. |
Navigating Limitations from Aqueous Boundary Layers and Paracellular Transport
Troubleshooting Guide: FAQs on ABLs and Paracellular Transport in Caco-2 Models
This guide addresses common experimental challenges and provides targeted solutions to improve the accuracy of your Caco-2 permeability assessments, particularly within research focused on optimizing permeability through molecular pair analysis.
FAQ 1: Why does our calculated active efflux not match our functional transport data, and how do ABLs influence this?
The Efflux Ratio (ER) is a standard metric to identify substrates of efflux transporters like P-glycoprotein (P-gp). A common pitfall is calculating the ER without accounting for the additional transport resistance from Aqueous Boundary Layers (ABLs), which can lead to significant underestimation of active transport [44].
- The Problem: Traditional models expressing ER as a function of only intrinsic membrane permeability (P₀) and efflux transporter permeability (P₵ₚᵩ) can be inaccurate. Single flux measurements in either the apical-to-basolateral (A-B) or basolateral-to-apical (B-A) direction are heavily affected by the additional resistance of ABLs and the filter support [44].
- The Solution: Incorporate the resistances of ABLs and the filter into your permeability model. Research confirms that a more comprehensive model which includes these factors provides more accurate access to the intrinsic P₵ₚᵩ value, leading to a better quantitative understanding of efflux [44].
FAQ 2: Can paracellular transport mask the detection of active efflux in our Caco-2 assays?
Yes, dominant paracellular transport can obscure active efflux, potentially leading to false negatives in your screening data [44].
- The Problem: The paracellular pathway facilitates the permeation of hydrophilic, low molecular weight compounds via the tight junctions between enterocytes. When this pathway is the dominant route of transport, it can reduce the observed Efflux Ratio to unity (ER ≈ 1), effectively masking any concurrent active efflux processes [44].
- The Solution:
- Characterize the Pathway: Use specific hydrophilic compounds that are known to traverse the paracellular route (e.g., mannitol, urea) to quantify the contribution of this pathway in your experiments [45].
- Experimental Control: Ensure your Caco-2 monolayers have formed tight junctions with high Transepithelial Electrical Resistance (TEER) values. A low TEER may indicate overly leaky junctions, causing paracellular dominance [46].
FAQ 3: How do we accurately quantify the impact of a new chemical entity on tight junction integrity?
Quantifying changes in the effective pore radius of tight junctions is key to understanding a compound's effect on the paracellular pathway.
- The Solution: Follow an established experimental and theoretical template that measures the permeability coefficients (P) of a series of hydrophilic compounds that vary in molecular size and charge (e.g., neutral mannitol, cationic atenolol, anionic lactate) [45].
- Factor out the mass transfer resistances of the filter support and ABLs to yield the true paracellular permeability coefficient (PP) [45].
- Analyze the changes in the permeation of these solutes using the Renkin molecular sieving function to calculate the effective aqueous pore radius of the tight junctions. This allows you to quantitatively assess the effect of perturbants on the paracellular pathway [45].
Experimental Protocols for Investigating Transport Pathways
Protocol 1: Differentiating Transport Routes in Caco-2 Monolayers
Objective: To delineate the contributions of passive transcellular, active efflux, and paracellular transport for a test compound.
Methodology:
- Cell Culture: Seed Caco-2 cells (e.g., HTB-37 from ATCC) on collagen-coated transwell filters at a density that optimizes confluent monolayer formation. Use culture medium such as DMEM supplemented with 10-20% FBS, 1% Non-Essential Amino Acids (NEAA), and 1% penicillin/streptomycin [5] [46].
- Validation of Monolayer Integrity: Monitor the formation of a tight barrier by measuring TEER regularly. Differentiated monolayers typically achieve TEER values above 300 Ω·cm² after 21 days in culture [46].
- Bidirectional Permeability Assay:
- A-B & B-A Transport: Measure the apparent permeability (Papp) of the test compound in both apical-to-basolateral and basolateral-to-apical directions.
- Efflux Transporter Inhibition: Repeat the bidirectional assay in the presence of a specific efflux transporter inhibitor (e.g., GF120918 for BCRP). A reduction in the B-A/A-B Papp ratio confirms the compound as an efflux transporter substrate [47].
- Paracellular Marker Co-Transport: Include a known paracellular marker like mannitol or urea in your A-B transport experiments. The permeability of your test compound relative to the marker helps estimate the paracellular contribution [45].
Protocol 2: Quantifying Tight Junction Perturbation
Objective: To determine the effect of a perturbant on the effective pore radius of tight junctions.
Methodology:
- Monolayer Preparation: Differentiate Caco-2 cells on transwell filters as described in Protocol 1.
- Perturbant Exposure: Treat the monolayers with the compound of interest (e.g., a permeation enhancer like palmitoyl-DL-carnitine) across a range of concentrations [45].
- Multicomponent Paracellular Flux: Measure the permeability of a panel of hydrophilic compounds that vary in size and charge (neutral, cationic, anionic) after perturbant exposure.
- Data Analysis:
Key Experimental Parameters for Paracellular Transport Studies
The table below summarizes quantitative data on the permeability of model compounds and the effects of perturbants, which can serve as benchmarks for your experiments [45].
Table 1: Paracellular Permeability and Perturbant Effects on Tight Junctions
| Compound / Perturbant | Key Finding / Permeability Value | Experimental Context |
|---|---|---|
| Mannitol (Neutral) | Used as a marker for molecular size-restricted diffusion. | Model compound for quantifying paracellular pathway activity. |
| Atenolol (Cationic) | Permeates cellular tight junctions faster than its neutral counterpart. | Demonstrates the influence of charge on paracellular diffusion. |
| Lactate (Anionic) | Permeates cellular tight junctions slower than its neutral counterpart. | Demonstrates the influence of charge on paracellular diffusion. |
| EGTA (Perturbant) | Causes a dramatic opening of TJs over a narrow concentration range (1.35-1.4 mM). | Ca++-dependent mechanism; used to experimentally modulate tight junctions. |
| Palmitoyl-DL-carnitine (Perturbant) | Produces a dose-dependent response in pore size (0 to 0.15 mM), plateauing at >0.15 mM. | Ca++-independent mechanism; used to experimentally modulate tight junctions. |
| Effective Pore Radius | Can be analyzed from 4.6 to 14.6 Å in effective radius using the Renkin function. | Quantitative measure of tight junction status after perturbation. |
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 2: Key Research Reagent Solutions for Caco-2 Transport Studies
| Item | Function / Application in Research |
|---|---|
| Caco-2 Cells | Human colorectal adenocarcinoma cell line; spontaneously differentiates into enterocyte-like cells, forming polarized monolayers with tight junctions. |
| Transwell Filters | Permeable supports for growing cell monolayers, allowing separate access to apical and basolateral compartments for permeability assays. |
| DMEM / MEM Medium | Base culture media; DMEM is commonly used and requires supplementation with FBS (10-20%) and NEAA for optimal Caco-2 growth [5] [46]. |
| Non-Essential Amino Acids (NEAA) | Crucial medium supplement; omission can lead to decreased Caco-2 growth rate and increased floating cells [5]. |
| Fetal Bovine Serum (FBS) | Standard serum supplement; typically used at 20% concentration for Caco-2 cultures to promote cell adhesion and growth [5]. |
| Efflux Transporter Inhibitors | Pharmacological tools (e.g., Elacridar/GF120918 for BCRP, Cyclosporine for P-gp) to confirm transporter involvement in compound efflux [47]. |
| Paracellular Markers | Hydrophilic compounds (e.g., Mannitol, Urea, Atenolol) used to probe the integrity and characteristics of the paracellular pathway [45]. |
| Tight Junction Perturbants | Agents (e.g., EGTA, Palmitoyl-DL-carnitine) used to experimentally and reversibly modulate the opening of tight junctions for mechanistic studies [45]. |
Visualizing Experimental Workflows and Transport Mechanisms
Caco-2 Permeability Assessment Workflow
Intestinal Drug Transport Mechanisms
Within the framework of a broader thesis on optimizing Caco-2 permeability through molecular pair analysis research, the implementation of high-throughput experimental techniques is paramount. The traditional Caco-2 permeability assay, while being the "gold standard" for predicting human intestinal absorption, is plagued by a very low throughput. The standard protocol requires at least 21 days of cell culture to establish a fully differentiated monolayer, which is then used for a single permeability assay during its stable period (up to day 30) [6] [48]. This bottleneck severely limits the pace of drug discovery and development. Molecular pair analysis research, which systematically explores the effects of small structural changes on permeability, necessitates the screening of numerous analogous compounds. Therefore, validated strategies to increase experimental throughput are essential. This technical support center document details a validated protocol for the re-use of Caco-2 monolayers, a method that can triple the throughput of this critical assay while maintaining data integrity, directly supporting the efficient generation of robust permeability data for in silico model development [6] [10] [16].
Validated Experimental Protocol for Monolayer Re-use
The following section provides a detailed, step-by-step methodology for the re-use of Caco-2 monolayers in permeability assays, as validated by extensive research [6] [48].
Materials and Pre-Assay Preparation
- Cell Line and Culture: Caco-2 cells (e.g., ECACC 09042001), passages 95-105, cultured in DMEM high glucose supplemented with 10% FBS and 1% non-essential amino acids [48].
- Seeding: Seed cells at a density of 2.6 × 10^5 cells/cm² on 12-well Transwell polycarbonate membrane inserts (1.12 cm² surface area, 0.4 µm pore size) [6] [48].
- Differentiation: Maintain cells for 21 days to achieve full differentiation, renewing medium every 2-3 days. The monolayers are stable for experiments from day 21 to day 30 post-seeding [6].
Integrity Assessment and Permeability Assay Workflow
The core of the re-use protocol hinges on a rigorous integrity check before each permeability experiment. The workflow for the initial and subsequent re-use assays is as follows:
Pre-Assay Integrity Check (Before any permeability test):
- TEER Measurement: Wash monolayers twice with pre-warmed HBSS-HEPES buffer (pH 7.4). Measure the Transepithelial Electrical Resistance (TEER) using a voltohmeter. Only use monolayers with TEER values indicating confluent and tight barriers (e.g., >500 Ω·cm²) [6].
- Paracellular Marker Flux: Perform a permeability assay using a paracellular marker like Lucifer Yellow (LY). A commonly used concentration is 100 µM. Sample from the basolateral side at 60 minutes. Calculate the apparent permeability (Papp). Monolayers with Papp(LY) ≤ 2.0 × 10⁻⁶ cm/s are considered intact and suitable for drug permeability assays [6].
Permeability Assay Execution:
- Dosing: Add the test compound dissolved in HBSS-HEPES buffer to the donor compartment (e.g., apical for A-B transport). The receiver compartment contains blank buffer.
- Incubation: Place the plate on an orbital shaker (50 rpm) in an incubator at 37°C without CO₂.
- Sampling: At predetermined time points (e.g., 10, 20, 30, 60 min), sample from the receiver compartment and replace with fresh buffer.
- Analysis: Quantify the compound concentration in the samples using a suitable analytical method (e.g., HPLC, fluorescence spectrometry).
Post-Assay Recovery and Validation for Re-use
- Post-Assay Wash: Following the permeability assay, carefully decant the dosing solutions.
- Recovery Incubation: Add fresh, pre-warmed complete culture media to both the apical and basolateral sides of the monolayer. Return the plate to the standard cell culture incubator (37°C, 5% CO₂) for a full two-day recovery period [6] [49]. Note: A one-day incubation is insufficient for full recovery of monolayer integrity.
- Re-validation for Re-use: After the two-day recovery, perform the Pre-Assay Integrity Check (TEER and LY Papp) again. The monolayer is ready for re-use only if the TEER has returned to its pre-assay value and the LY Papp is within the acceptable limit [6].
Using this protocol, a single Caco-2 monolayer can be reliably used for permeability assays on days 22, 25, and 28 post-seeding, effectively tripling the throughput [6] [48].
Troubleshooting Guide & FAQs
This section addresses specific, frequently encountered issues when implementing the monolayer re-use protocol.
Frequently Asked Questions
| Question | Answer & Solution |
|---|---|
| Can all types of transport mechanisms be studied with re-used monolayers? | The protocol is fully validated for compounds that permeate via passive transcellular and paracellular routes [48]. Preliminary data for carrier-mediated transport (e.g., P-gp efflux, SGLT1 influx) is promising, but requires further investigation and lab-specific validation before implementation for such compounds [48]. |
| The TEER does not recover after the two-day incubation. What could be wrong? | This indicates monolayer stress or damage. Potential causes: (1) Toxic test compounds: Pre-evaluate compound cytotoxicity using an MTT assay [6]. (2) Physical damage during handling: Use careful pipetting techniques. (3) Microbial contamination: Check media for sterility. If TEER does not recover, discard the monolayer. |
| Why is a two-day recovery necessary? Why not one day? | Research has shown that the permeability assay causes a small but significant decrease in TEER. A one-day incubation is insufficient for full recovery. A two-day incubation with culture media is required and sufficient for the TEER to return to its original value, indicating the re-establishment of tight junctions [6] [49]. |
| The Papp values for my control compounds are inconsistent between the first and second use. | Minor variations are normal. Ensure the integrity parameters (TEER and LY Papp) are nearly identical before each assay. If large discrepancies occur, verify your sampling and analytical techniques. Inconsistencies may also arise if the test compounds from the first assay were not thoroughly washed out. |
| Are there more modern methods to monitor integrity in real-time? | Yes. Impedance-based real-time cell analyzers (e.g., xCELLigence RTCA) can non-invasively monitor monolayer integrity, growth, and quality continuously, providing more robust data than endpoint TEER measurements [20]. |
Research Reagent Solutions
The table below lists the key materials and reagents essential for successfully implementing the Caco-2 monolayer re-use protocol.
Table 1: Essential Reagents and Materials for the Caco-2 Re-use Protocol
| Item | Function & Role in the Protocol | Example & Specification |
|---|---|---|
| Caco-2 Cell Line | The human colonic adenocarcinoma cell line that, upon differentiation, forms a polarized intestinal epithelial monolayer. | ECACC 09042001, passages 95-105 [6] [48]. |
| Transwell Inserts | Semi-permeable membrane supports that allow cell polarization and permeability measurements. | 12-well plate, Polycarbonate membrane, 1.12 cm² surface area, 0.4 µm pore size [6] [50]. |
| Culture Medium | Supports cell growth and differentiation, and is critical for the 2-day post-assay recovery. | DMEM high glucose, 10% FBS, 1% Non-Essential Amino Acids, 1% Pen/Strep [48]. |
| Transport Buffer | The physiologically-compatible buffer used during the permeability assay. | HBSS supplemented with 25 mM HEPES, pH 7.4 [6] [48]. |
| TEER Voltohmeter | Device to measure Transepithelial Electrical Resistance, the primary metric for monolayer integrity. | e.g., EVOM2 Voltohmeter with "chopstick" electrodes [6]. |
| Integrity Marker (Lucifer Yellow) | A paracellular pathway marker used to quantitatively validate monolayer tightness before each assay. | Lucifer Yellow CH di-potassium salt; used at 100 µM [6] [49]. |
| Orbital Shaker | Provides gentle agitation during the permeability assay to minimize the unstirred water layer effect. | IKA-Schüttler MTS4 or equivalent, set to 50 rpm [48]. |
Data Presentation & Validation
The following tables summarize the key quantitative data that validates the re-use protocol, providing a reference for researchers to compare their own results against.
Table 2: Throughput Comparison: Standard vs. Re-use Protocol
| Protocol Feature | Standard Protocol | Re-use Protocol (Proposed) | Throughput Gain |
|---|---|---|---|
| Culture Period | 21-30 days | 21-30 days | Same |
| Permeability Assays per Monolayer | 1 | 3 (e.g., on days 22, 25, 28) | 3-fold increase [6] [48] |
| Resource Consumption | High (1 insert per compound) | Reduced (1 insert per 3 compounds) | ~66% reduction in inserts, cells, and media |
Table 3: Validation Data for Monolayer Integrity Upon Re-use
| Integrity Parameter | Initial Assay (Day 22) | First Re-use (Day 25) | Second Re-use (Day 28) | Validation Criterion |
|---|---|---|---|---|
| TEER Value (Ω·cm²) | Pre-assay: e.g., 650 ± 50 | Recovers to pre-assay value (e.g., 645 ± 45) | Recovers to pre-assay value (e.g., 640 ± 55) | Full recovery after 2-day incubation [6] |
| LY Papp (×10⁻⁶ cm/s) | ≤ 2.0 | ≤ 2.0 | ≤ 2.0 | No significant increase [6] |
| Tight Junction Staining (ZO-1) | Continuous, well-defined | Continuous, well-defined | Continuous, well-defined | Morphological confirmation [6] |
Conceptual Framework: Integrating Re-use Protocols with Molecular Pair Analysis
The development and validation of this re-use protocol is not an isolated effort. It is a critical enabler for the broader research goal of understanding and predicting Caco-2 permeability through molecular pair analysis (MMPA). The relationship between these components is illustrated below and forms the conceptual backbone of the thesis.
This framework demonstrates that the experimental protocol directly feeds high-quality, volume data into computational workflows. Machine learning models (e.g., XGBoost, Random Forest) trained on such datasets can achieve high accuracy in predicting Caco-2 permeability [10] [16]. Subsequent Matched Molecular Pair Analysis then allows researchers to derive clear, interpretable rules on how specific structural changes (e.g., adding a methyl group, changing a halogen) affect permeability, moving from black-box prediction to actionable design guidance [10] [16]. This virtuous cycle of experimental optimization, data generation, and computational modeling accelerates the entire drug discovery pipeline.
Frequently Asked Questions
FAQ 1: Why is dataset balancing a critical pre-processing step specifically for Caco-2 permeability multiclass modeling?
In multiclass Caco-2 permeability modeling, the dataset is often imbalanced, meaning the number of molecules in each permeability category (e.g., high, medium, low) is not equal [51]. This class imbalance poses a significant challenge for developing predictive models, as machine learning algorithms may become biased toward the majority class [52] [53]. For instance, a model might achieve high accuracy by simply always predicting the most common class, but it would fail to accurately identify molecules with low or medium permeability, which are often critically important in drug discovery [53]. Employing balancing strategies ensures the model pays adequate attention to all permeability classes, leading to more reliable and robust predictions across the entire chemical space of interest [51].
FAQ 2: What are the primary data-level methods to balance an imbalanced Caco-2 permeability dataset?
The main data-level methods involve resampling the training data to create a more balanced class distribution [54]. These are implemented before model training and include:
- Oversampling: This method increases the number of instances in the minority class(s). Techniques range from simple random duplication to more advanced methods like the Synthetic Minority Oversampling Technique (SMOTE), which creates synthetic examples in the feature space rather than just copying [52] [53]. For example, one study applied ADASYN, an adaptive oversampling method, to achieve strong performance in multiclass Caco-2 permeability classification [51].
- Undersampling: This method reduces the number of instances in the majority class(s) by randomly removing samples until balance is achieved [54] [52]. While it can be efficient, it risks discarding potentially useful data.
Table 1: Comparison of Data-Level Balancing Methods for Caco-2 Modeling
| Method | Description | Advantages | Disadvantages | Reported Performance (Example) |
|---|---|---|---|---|
| Random Oversampling | Randomly duplicates existing minority class samples. | Simple to implement; no loss of information from original dataset. | Can lead to overfitting, especially if copies are identical. | Varies by dataset and base classifier. |
| SMOTE | Creates synthetic minority class samples by interpolating between existing ones [52]. | Reduces risk of overfitting compared to random oversampling; increases diversity. | May generate noisy samples if the minority class is not well clustered. | Improved validation accuracy from 90% to 94% in a text classification benchmark [53]. |
| Random Undersampling | Randomly removes samples from the majority class. | Reduces computational cost and training time. | Potentially discards useful, important data. | Accuracy: 0.727, Precision: 0.824, Logloss: 0.728 on a multi-class task [54]. |
| ADASYN | An adaptive oversampling method that generates more synthetic data for minority class examples that are harder to learn. | Focuses on the most difficult minority class cases. | Can amplify noise from the minority class. | Achieved test accuracy of 0.717 and MCC of 0.512 for multiclass Caco-2 prediction [51]. |
FAQ 3: Which algorithm-level methods can improve model performance on imbalanced Caco-2 data?
Instead of modifying the data, algorithm-level methods adjust the learning process itself to be more sensitive to minority classes. Key approaches include:
- Using Boosting Algorithms: Algorithms like XGBoost and CatBoost are ensemble methods that sequentially train models, with each new model focusing on correcting the errors of the previous ones [54]. This makes them naturally adept at handling imbalanced datasets. Studies have shown that XGBoost often provides superior predictions for Caco-2 permeability tasks [10]. Hyperparameter tuning of these models can further optimize performance metrics like log loss [54].
- Cost-Sensitive Learning: This involves assigning a higher misclassification cost to the minority classes during model training. Many algorithms allow you to set the
class_weightparameter to "balanced," which automatically adjusts weights inversely proportional to class frequencies [53]. This penalizes the model more for mistakes on the minority classes. - Specialized Ensemble Methods: The BalancedBaggingClassifier is an extension of traditional bagging that incorporates additional balancing during the training of each base estimator in the ensemble, such as resampling [52].
Table 2: Performance of Algorithm-Level Methods on a Multi-class Imbalanced Task
| Algorithm | Key Hyperparameters | Accuracy | Log Loss | Notes |
|---|---|---|---|---|
| CatBoost (Default) | Default parameters | 0.834 | 0.458 | Strong performance out-of-the-box [54]. |
| CatBoost (Tuned) | nestimators: 666, learningrate: 0.067, max_depth: 3 | - | 0.439 | Hyperparameter tuning with Optuna reduced log loss [54]. |
| XGBoost (Default) | Default parameters | 0.830 | 0.515 | Generally provides better predictions than comparable models for Caco-2 permeability [10]. |
| XGBoost (Tuned) | nestimators: 592, learningrate: 0.030, max_depth: 8 | - | 0.422 | Optimized XGBoost achieved the best log loss in this example [54]. |
FAQ 4: How should we evaluate model performance on a balanced but originally imbalanced Caco-2 dataset?
Accuracy can be a misleading metric for imbalanced datasets [52] [53]. A comprehensive evaluation should include:
- Confusion Matrix: Provides a detailed breakdown of correct and incorrect classifications for each class, revealing if the model is consistently failing on a specific permeability category [53].
- Precision, Recall, and F1-Score: It is crucial to examine these metrics for each class individually (e.g., macro or weighted averages) [52]. Precision measures the model's reliability when it predicts a specific class, while Recall measures its ability to find all samples of a class. The F1-score is the harmonic mean of precision and recall and is a preferred metric for imbalanced datasets [52].
- Matthews Correlation Coefficient (MCC): This is a more robust measure that considers true and false positives and negatives and is well-suited for imbalanced data, as reported in a Caco-2 multiclass study [51].
- Log Loss: This assesses the quality of the model's probability estimates, with a lower value indicating more confident and correct predictions [54].
FAQ 5: How can molecular pair analysis be integrated with dataset balancing strategies?
Molecular Pair Analysis (MPA) can be a powerful complement to balancing strategies. Once a reliable model is built on a balanced dataset, MPA can be used to extract chemical transformation rules that favorably impact Caco-2 permeability [10]. For example, a model with high interpretability can help identify key molecular descriptors. MPA can then analyze pairs of molecules that differ only by a specific substructure, quantifying how that change affects the permeability class. These rules can then guide medicinal chemists in optimizing lead compounds, for instance, by suggesting a specific functional group change that is likely to shift a molecule from "low" to "medium" permeability without altering other desired properties [10].
Experimental Protocols for Dataset Balancing
Protocol 1: Implementing SMOTE for Caco-2 Data Oversampling
This protocol details the use of SMOTE to balance a Caco-2 permeability dataset using Python.
Prerequisites: Install the necessary libraries:
imbalanced-learn(imblearn),scikit-learn, andpandas.Load and Split Data: Load your Caco-2 dataset, where
Xcontains the molecular descriptors/fingerprints andycontains the permeability classes (e.g., 0, 1, 2 for Low, Medium, High). Split into training and test sets. Crucially, apply resampling only to the training data to avoid data leakage.Apply SMOTE: Use the
SMOTEclass from imblearn to oversample the minority classes in the training set.Verify and Train: Check the new class distribution and proceed to train your chosen classifier on the balanced training data (
X_train_resampled,y_train_resampled). Evaluate on the untouched test set (X_test,y_test).
Protocol 2: Hyperparameter Tuning for XGBoost on a Balanced Dataset
This protocol uses Optuna to optimize XGBoost hyperparameters for a multi-class classification task.
Prepare Data: Ensure your training data is balanced using one of the methods above (e.g., SMOTE). Define your features (
X_train_bal) and labels (y_train_bal).Define the Objective Function: This function defines the hyperparameter space and the goal (minimizing log loss).
Run the Optimization Study: Execute the Optuna study to find the best hyperparameters.
Workflow Visualization
Multiclass Permeability Modeling Workflow
Balancing Strategy Decision Logic
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Tools for Caco-2 Permeability Modeling & Data Balancing
| Tool / Reagent | Function in Context | Technical Notes |
|---|---|---|
| Caco-2 Cell Line | In vitro model of the human intestinal mucosa used to generate experimental permeability data [3] [55]. | Watch for passage number-induced genomic instability and phenotypic drift; limit continuous cultures [3]. |
| KNIME Analytics Platform | An open-source platform for building automated workflows for data blending, curation, QSPR modeling, and visualization [55]. | Enables creation of reproducible workflows for data cleaning, feature selection, and consensus model building. |
| Imbalanced-learn (imblearn) | A Python toolbox specifically for tackling dataset imbalance [54] [52]. | Provides implementations of SMOTE, ADASYN, RandomOverSampler, RandomUnderSampler, and ensemble variants. |
| XGBoost / CatBoost | High-performance gradient boosting frameworks designed for efficiency and model performance [54] [10]. | Often provide superior predictions for Caco-2 tasks. Support native handling of categorical data (CatBoost) and cost-sensitive learning. |
| Optuna | A hyperparameter optimization framework for automating the search for the best model parameters [54]. | Uses efficient algorithms like TPE to minimize a defined objective (e.g., log loss) over multiple trials. |
| RDKit | An open-source cheminformatics toolkit for calculating molecular descriptors and fingerprints [55]. | Used to transform molecular structures into numerical features (e.g., MOE-type descriptors, Morgan fingerprints) for ML models. |
Benchmarking Success: Validating MMPA Against Advanced Models and Industrial Data
FAQs: Core Concepts and Method Selection
Q1: What are the fundamental differences between MMPA and ML/DL for Caco-2 permeability prediction?
A1: Matched Molecular Pair Analysis (MMPA) and Machine Learning/Deep Learning (ML/DL) serve distinct but complementary roles.
- MMPA is a rule-based approach that identifies the effects of small, specific chemical transformations on a property like Caco-2 permeability. It provides clear, interpretable, and actionable design guidelines (e.g., "replacing a chlorine atom with a methyl group in this scaffold typically increases permeability") [16].
- ML/DL models are data-driven approaches that learn complex, non-linear relationships between a molecule's overall structure and its permeability. They are powerful for high-throughput prediction and screening of new compounds but often function as "black boxes," offering less direct insight for chemical optimization [16] [56].
Q2: When should I prioritize MMPA over ML/DL in my research?
A2: Prioritize MMPA when your goal is lead optimization. If you have a core scaffold and need to understand how specific structural changes will impact permeability, MMPA delivers direct, interpretable chemical transformation rules [16]. It is most effective when you have a series of structurally related analogs.
Q3: Our ML model for Caco-2 permeability performs well on public data but poorly on our internal compounds. What could be the cause?
A3: This is a common challenge due to the high experimental variability of Caco-2 assays across laboratories [55] [23]. Differences in cell culture conditions, passage number, monolayer age, and assay protocols can lead to systematic shifts in data. This affects model transferability. To address this:
- Use Assay-Ready Cells: Consider using pre-qualified, ready-to-use Caco-2 monolayers (e.g., CacoReady) to reduce internal variability [18].
- Fine-Tune Models: Retrain or fine-tune your public model on a small set of high-quality internal data to calibrate it to your specific assay conditions [16] [23].
- Employ Local Models: For project-specific chemical series, building a local model using only structurally similar compounds can sometimes outperform a global model [23].
Q4: How can I assess the reliability of a Caco-2 permeability prediction from an ML model?
A4: Two key concepts are Applicability Domain (AD) analysis and confidence metrics.
- Applicability Domain: Determine if the new compound you are predicting is structurally similar to the compounds used to train the model. Predictions for molecules outside the model's AD are less reliable [16] [55].
- Experimental Variability: Check if the model accounts for experimental noise. Some advanced models are trained on data curated by calculating the mean and standard deviation of repeated measurements, retaining only compounds with low variability (e.g., STD ≤ 0.3) for the final training set [16] [55].
Troubleshooting Guides
Troubleshooting MMPA
| Issue | Possible Cause | Solution |
|---|---|---|
| No meaningful chemical transformations are found. | The dataset lacks sufficient structural analogues or is too diverse. | Curate a dataset focused on a specific chemical series or scaffold from your internal medicinal chemistry programs. |
| Extracted transformation rules are contradictory. | The effect of a transformation is context-dependent (varies by chemical scaffold). | Segment the analysis by core scaffold or use context-aware MMPA. Do not apply rules universally without verification. |
| Rules from public data do not apply to your compounds. | The public dataset's chemical space or assay conditions differ significantly from your internal context. | Generate MMPA rules directly from your high-quality, internally generated Caco-2 permeability data [16]. |
Troubleshooting ML/DL Models
| Issue | Possible Cause | Solution |
|---|---|---|
| Poor model performance on external validation sets. | High experimental variability in training data; model overfitting. | Implement rigorous data curation: remove duplicates with high standard deviation, apply data cleaning workflows in platforms like KNIME, and use Y-randomization testing to validate robustness [16] [55]. |
| Model is a "black box" with no design insights. | Using complex models (e.g., deep neural networks) without interpretation tools. | Use models that offer feature importance (e.g., Random Forest) or combine global ML models with local MMPA to gain both predictive power and design insights [16] [31]. |
| Low predictive accuracy for specific permeability ranges. | Imbalanced dataset with few low-permeability compounds. | Apply targeted sampling or data augmentation techniques for under-represented classes. For e/bRo5 compounds, a local similarity-based model may be more effective than a global model [23]. |
Quantitative Data Comparison
Performance of Machine Learning Algorithms for Caco-2 Permeability Prediction
Table: Benchmarking ML Algorithms on Public Caco-2 Datasets (LogPapp Prediction)
| Algorithm | Molecular Representation | Test Set RMSE | Test Set R² | Key Advantages |
|---|---|---|---|---|
| XGBoost [16] | Morgan FP + RDKit 2D Descriptors | ~0.31 (on specific dataset) [16] | ~0.81 (on specific dataset) [16] | Generally provided better predictions than comparable models; handles diverse features well. |
| SVM-RF-GBM Ensemble [31] | Selected Molecular Descriptors | 0.38 | 0.76 | Superior performance by leveraging strengths of multiple algorithms. |
| Random Forest (KNIME) [55] | Morgan FP + Physicochemical Descriptors | 0.43 - 0.51 | 0.57 - 0.61 | Produces interpretable models; suitable for automated workflows. |
| LightGBM [23] | RDKit Descriptors | Not explicitly stated | Not explicitly stated | Highly efficient and suitable for large-scale screening; identified as a top performer. |
| Support Vector Machine (SVM) [31] | Selected Molecular Descriptors | 0.39 - 0.40 | 0.73 - 0.74 | Good performance for non-linear relationships. |
In Vitro to In Vivo Correlation for Caco-2 Assay
Table: Interpreting Caco-2 Papp Values for Predicting Human Intestinal Absorption [18]
| In Vitro Papp (cm/s) | Predicted In Vivo Absorption | Interpretation for Drug Development |
|---|---|---|
| ≤ 1.0 × 10⁻⁶ | Low (0-20%) | High risk for poor oral bioavailability; may require structural modification or alternative delivery. |
| 1.0 × 10⁻⁶ to 10 × 10⁻⁶ | Medium (20-70%) | Moderate absorption; candidate for further optimization to improve permeability. |
| > 10 × 10⁻⁶ | High (70-100%) | Favorable permeability; unlikely to be the limiting factor for oral absorption. |
Experimental Protocols
Standard Protocol for Caco-2 Permeability Assay
Key Materials:
- Caco-2 Cells: Human colon adenocarcinoma cells (e.g., CacoReady plates [18] or cultured cells [5]).
- Growth Medium: MEM or DMEM, supplemented with 20% Fetal Bovine Serum (FBS), 1% Non-Essential Amino Acids (NEAA), and 1% Penicillin/Streptomycin [5] [18].
- Transwell Inserts: 24-well or 96-well plates with porous polyester filters [18].
- Transport Buffer: Hanks' Balanced Salt Solution (HBSS) or similar, pH 7.4.
- Control Compounds: Propranolol (high permeability), Atenolol (low permeability), Digoxin (P-gp substrate) [18].
Procedure:
- Cell Culture and Seeding: Seed Caco-2 cells onto Transwell inserts at a high density. Culture for 15-21 days, with medium changes every 48-72 hours, to allow formation of a confluent and differentiated monolayer [18].
- Monolayer Integrity Check: Before the experiment, measure Transepithelial Electrical Resistance (TEER). Acceptable values are >1000 Ω·cm² for 24-well plates and >500 Ω·cm² for 96-well plates. Validate integrity using a paracellular marker like Lucifer Yellow (Papp ≤ 1 × 10⁻⁶ cm/s) [18].
- Permeability Assay:
- Add the test compound (e.g., 10 µM suggested for unknowns) to the donor compartment (apical for A-B, basal for B-A).
- Incubate at 37°C for a set time (e.g., 2 hours). Sample from the receiver compartment at the end of the incubation [18].
- Sample Analysis: Quantify compound concentration in the samples using a sensitive method like LC-MS/MS [18].
- Data Calculation: Calculate the Apparent Permeability (Papp) using the formula: Papp (cm/s) = (dQ/dt) / (A × C₀) where dQ/dt is the transport rate (nmol/s), A is the membrane surface area (cm²), and C₀ is the initial donor concentration (nmol/mL) [18].
Protocol for Developing a Machine Learning QSPR Model
Key Materials:
- Software: KNIME Analytics Platform, Python with scikit-learn, RDKit, LightGBM, or other ML libraries [55] [23].
- Dataset: A curated dataset of chemical structures (SMILES) and corresponding experimental LogPapp values [16] [55].
Procedure:
- Data Curation and Standardization:
- Collect data from public and internal sources [16] [55].
- Standardize molecular structures using RDKit's MolStandardize module to achieve consistent tautomer and neutral forms [16] [23].
- Handle duplicates by calculating the mean and standard deviation of replicate measurements. Retain only entries with low variability (e.g., STD ≤ 0.3) for model training [16] [55].
- Molecular Representation:
- Feature Selection:
- Model Training and Validation:
- Split data into training, validation, and test sets (e.g., 8:1:1 ratio) [16].
- Train multiple algorithms (e.g., XGBoost, RF, SVM, GBM) and use cross-validation to tune hyperparameters.
- Validate the final model on a held-out test set and an external test set (e.g., in-house data) to evaluate its predictive power and generalizability [16] [23].
Research Reagent Solutions
Table: Essential Materials for Caco-2 Permeability and Modeling Workflows
| Item | Function/Application | Example/Specification |
|---|---|---|
| Ready-to-Use Caco-2 Monolayers | Saves cell culture time, ensures consistent monolayer quality and integrity for assays. | CacoReady plates (24-well & 96-well formats) [18]. |
| Caco-2 Cell Culture Medium | Supports optimal growth and spontaneous differentiation of Caco-2 cells into enterocyte-like cells. | MEM or DMEM, 20% FBS, 1% NEAA, 1% P/S [5]. |
| Reference Compounds | Validate assay performance by confirming expected permeability and transporter activity. | Propranolol (High Perm), Atenolol (Low Perm), Digoxin (P-gp substrate) [18]. |
| KNIME Analytics Platform | Open-source platform for building automated, end-to-end workflows for data curation, QSPR modeling, and prediction. | Includes nodes for RDKit descriptor calculation, data preprocessing, and machine learning [55]. |
| RDKit | Open-source cheminformatics toolkit for calculating molecular descriptors, fingerprints, and standardizing structures. | Essential for preparing molecular representations for ML models [16] [55]. |
Methodology Visualization
Integrated Workflow for Caco-2 Permeability Optimization
Machine Learning Model Development and Validation Process
A pressing challenge in modern drug discovery is the performance drop of predictive models when applied outside their original training environment. Models developed on public research data, such as those for predicting Caco-2 permeability, often face a "generalization gap" when deployed on proprietary pharmaceutical R&D datasets [57] [16]. This discrepancy arises from differences in experimental protocols, measurement techniques, and population biases between public and private data sources. This technical support center provides troubleshooting guides and experimental protocols to help researchers diagnose, address, and overcome these transferability issues, with a specific focus on optimizing Caco-2 permeability through molecular pair analysis.
Troubleshooting Guides: Diagnosing Transferability Failures
Guide: Diagnosing Poor Model Performance on Internal Data
Observed Problem: A Caco-2 permeability model trained on public data shows significantly degraded performance (e.g., >20% increase in RMSE) when predicting on your internal compound library.
Investigation and Resolution:
Step 1: Verify Data Distribution Shifts
- Action: Compare the distributions of key molecular descriptors (e.g., Molecular Weight, LogP, HBD, HBA) between your public training set and internal dataset.
- Tool: Use Principal Component Analysis (PCA) or t-SNE plots to visualize structural differences.
- Interpretation: A significant overlap in the PCA plot suggests other issues; distinct clusters indicate your internal compounds may lie outside the model's applicability domain.
Step 2: Assess Applicability Domain (AD)
- Action: Calculate the distance of your internal compounds to the public training set used for the model.
- Tool: Use leverage-based methods or distance-to-model metrics (e.g., using Euclidean distance in the descriptor space).
- Interpretation: If a large proportion (>15%) of your internal compounds fall outside the pre-defined AD, the model is being applied extrapolatively and its predictions are unreliable [16].
Step 3: Check for Systematic Measurement Bias
- Action: Select a small, diverse set of 20-30 compounds from your internal library. Compare their experimental Papp values with published values from the public data source for the same compounds.
- Interpretation: A consistent, non-random offset (e.g., your values are systematically lower) indicates a lab-specific bias. This may require model calibration or adjustment.
Solution Path:
- If AD is the issue: Retrain the model by incorporating a broader public dataset or a representative subset of your internal data to expand its chemical space knowledge.
- If bias is the issue: Apply a simple linear correction based on your comparative analysis, or use Transfer Learning techniques to fine-tune the public model on a small portion of your high-quality internal data.
Guide: Handling Inconsistent Predictions for Structural Analogs
Observed Problem: Your model predicts drastically different permeability values for chemically similar compounds, contrary to experimental evidence.
Investigation and Resolution:
Step 1: Interrogate Model Interpretability
- Action: Use SHAP (SHapley Additive exPlanations) or similar techniques on the problematic compounds to identify which molecular features the model is over- or under-weighting.
- Interpretation: The model might be latching onto irrelevant, spurious correlations from the public dataset that do not hold in your specific chemical series.
Step 2: Perform Matched Molecular Pair Analysis (MMPA)
- Action: Systematically identify the specific chemical transformation (e.g., -OH to -OCH3) that is causing the prediction discontinuity.
- Tool: Use the RDKit or KNIME platforms to execute MMPA.
- Interpretation: MMPA can reveal if the model fails to correctly learn the effect of a common transformation. This provides a concrete "rule" for model correction and offers direct insights for chemical optimization [16].
Solution Path:
- Feature Refinement: Retrain the model using a different set of molecular descriptors or fingerprints that better capture the relevant physicochemical properties for permeability (e.g., those related to hydrogen bonding).
- Data Augmentation: Curate additional public data or generate synthetic data points that specifically exemplify the correct permeability relationship for the problematic transformation.
Frequently Asked Questions (FAQs)
Q1: Which machine learning algorithm generalizes best for Caco-2 prediction when moving from public to industrial data?
A: Based on comprehensive benchmarking, tree-based ensemble methods, particularly XGBoost, have demonstrated superior and more robust transferability compared to other models like Random Forest (RF) and Support Vector Machine (SVM), and even some deep learning architectures like DMPNN [16]. XGBoost's regularization techniques help prevent overfitting to the noise and specific biases of public datasets, leading to better performance on external industrial data.
Q2: What is a typical performance drop we should expect when applying a public model to our in-house data?
A: The performance drop varies, but it can be substantial. Benchmarking studies on drug response prediction have shown that models can experience significant performance degradation when applied to unseen datasets from different sources [57]. For Caco-2 permeability, one industrial validation study reported that boosting models trained on public data "retained a degree of predictive efficacy" on an internal pharmaceutical dataset, but specific performance metrics like R² and RMSE were noticeably worse than on the original public test sets [16]. Always validate the public model's performance on a small, representative sample of your internal data before deployment.
Q3: How can we quickly evaluate if a public Caco-2 model is suitable for our specific project's chemical space?
A: The most effective method is to perform an Applicability Domain (AD) analysis [16]. This involves:
- Calculating the molecular descriptor profiles of your project's compound library.
- Comparing them to the descriptor profiles of the public model's training set.
- Quantifying how many of your compounds fall within the model's known chemical space. A high percentage (e.g., >85%) inside the AD suggests the model is likely to be reliable for your project.
Q4: Beyond retraining the model, what strategies can improve transferability?
A: Two effective strategies are:
- Model Calibration: Adjust the output of the pre-trained model using a small set of your internal experimental data (e.g., 20-50 compounds) to correct for systematic bias.
- Ensemble Modeling: Combine predictions from multiple models trained on different public datasets. The diversity of the ensemble can often lead to more robust predictions on new, external data [57].
Experimental Protocols for Validation and Improvement
Protocol: Industrial Validation of a Public Caco-2 Model
This protocol outlines the steps to rigorously test a publicly available Caco-2 permeability model on an internal pharmaceutical company dataset.
1. Objective: To evaluate the predictive performance and generalizability of a public Caco-2 permeability model on an internal compound library.
2. Materials and Reagents
- Internal Validation Set: 50-100 compounds from your corporate library with reliably measured Caco-2 Papp values (preferably measured using a standardized internal protocol).
- Software: The public model, implemented in a suitable environment (e.g., Python, R, KNIME).
- Computational Tools: Scripts for calculating molecular descriptors/fingerprints and for performing statistical analysis (e.g., R², RMSE, MAE).
3. Methodology 1. Data Curation: Standardize the molecular structures of your internal validation set (e.g., using RDKit). Ensure the permeability values are in the same unit (e.g., 10⁻⁶ cm/s) and scale (e.g., log10) as the public model's training data. 2. Descriptor Alignment: Generate the exact same type of molecular features (e.g., ECFP4 fingerprints, RDKit 2D descriptors) used by the public model for your internal compounds. 3. Blind Prediction: Use the public model to predict the Caco-2 permeability for every compound in your internal validation set. Do not train or fine-tune the model at this stage. 4. Performance Assessment: Calculate the correlation (R²/Pearson's r) and error metrics (RMSE, MAE) between the model's predictions and your experimental values. 5. Applicability Domain Analysis: Determine the percentage of your internal compounds that fall within the model's applicability domain.
4. Expected Output: A quantitative report detailing the model's performance on your internal data, identifying any systematic biases, and providing a list of compounds for which the model's predictions are considered unreliable (those outside the AD).
Protocol: Utilizing Matched Molecular Pair Analysis (MMPA) for Permeability Optimization
This protocol leverages MMPA to extract chemically meaningful insights from model predictions or experimental data to guide the optimization of Caco-2 permeability.
1. Objective: To identify specific chemical transformations that consistently lead to increased Caco-2 permeability, providing actionable guidance for medicinal chemistry.
2. Materials
- Dataset: A large and curated dataset of Caco-2 permeability measurements (public, internal, or combined).
- Software: KNIME, RDKit, or specialized MMPA software.
3. Methodology 1. Data Preparation: Input a dataset of molecules and their corresponding Caco-2 permeability values (experimental or predicted). 2. Pair Identification: The MMPA algorithm systematically breaks down each molecule into a constant core and a variable R-group, identifying all pairs of compounds that differ only by a single, well-defined structural transformation at a single site. 3. Delta Calculation: For each matched molecular pair (e.g., Compound A: -H, Compound B: -CH₃), calculate the difference in their permeability values (ΔPapp). 4. Rule Extraction: Aggregate the results for each unique transformation. A transformation that consistently leads to a positive ΔPapp across multiple different molecular contexts represents a robust rule for improving permeability. 5. Contextual Analysis: Investigate if the effect of a transformation is dependent on the local chemical environment (e.g., the effect of adding -Cl to an aromatic ring might differ if it's ortho to a hydrogen bond donor).
4. Expected Output: A ranked list of chemical transformations (e.g., "replacing a methyl ester with a primary amide typically decreases logPapp by 0.3-0.5 units") that can be directly used by medicinal chemists to prioritize synthetic efforts [16].
Visualization of Workflows
Model Transferability Assessment
The following diagram illustrates the logical workflow for assessing the transferability of a public model to an internal pharmaceutical R&D dataset.
Permeability Optimization via MMPA
This diagram outlines the process of using Matched Molecular Pair Analysis to derive actionable design rules from Caco-2 permeability data.
Data Presentation: Model Performance and Reagents
Table 1: Benchmarking Model Generalization for Caco-2 Permeability Prediction
This table summarizes findings from a study that evaluated the performance of various machine learning models on public data and their subsequent transferability to an industrial dataset [16].
| Model / Algorithm | Public Test Set Performance (Avg. RMSE) | Industrial Validation Set Performance (RMSE) | Relative Performance Drop |
|---|---|---|---|
| XGBoost | 0.41 | 0.51 | ~24% |
| Random Forest (RF) | 0.43 | 0.56 | ~30% |
| Support Vector Machine (SVM) | 0.48 | 0.65 | ~35% |
| Deep Neural Network (DMPNN) | 0.45 | 0.62 | ~38% |
| CombinedNet | 0.42 | 0.58 | ~38% |
Table 2: Research Reagent Solutions for Caco-2 & In Silico Modeling
This table details key materials and computational tools used in the field for both experimental and computational studies of permeability.
| Item Name | Type | Function / Application |
|---|---|---|
| Caco-2 Cell Line | Biological Model | Human colon adenocarcinoma cell line that differentiates to form an intestinal epithelial monolayer, serving as the gold standard for in vitro permeability assessment [9] [16]. |
| HDM-PAMPA | Assay Kit | High-Throughput Parallel Artificial Membrane Permeability Assay used to determine hexadecane/water partition coefficients (K_hex/w), which can accurately predict intrinsic Caco-2/MDCK permeability [58]. |
| RDKit | Software Tool | An open-source cheminformatics toolkit used for molecular standardization, descriptor calculation, fingerprint generation (e.g., Morgan fingerprints), and Matched Molecular Pair Analysis [16]. |
| ADMET Predictor | Software Module | A commercial tool that provides predictive models for ADMET properties, including classification models for transporters (e.g., Pgp, BCRP) and prediction of metabolic parameters [59]. |
| COSMOtherm | Software Tool | A physics-based software for predicting solvation thermodynamics and partition coefficients, which can be used as an in silico alternative to experimental K_hex/w measurements [58]. |
Correlating In Vitro Caco-2 Papp with Human Fraction Absorbed (fa)
Frequently Asked Questions (FAQs)
FAQ 1: How do I convert a Caco-2 Papp value into a prediction for human Fraction Absorbed (Fa)?
The apparent permeability coefficient (Papp) obtained from Caco-2 assays can be correlated to the human Fraction Absorbed (Fa) using established in vitro-in vivo correlation (IVIVC) models. The process involves a two-step calculation to first estimate human effective permeability (Peff) and then calculate Fa [60].
Step 1: Estimate Human Jejunal Permeability (Peff) The following equation describes the correlation between Caco-2 Papp and human Peff [60]:
log(Peff) = 0.4926 · log(Papp) – 0.1454Peff= Human effective permeability (10⁻⁴ cm/s)Papp= Apparent permeability from Caco-2 assay (10⁻⁶ cm/s)
Step 2: Predict Fraction Absorbed (Fa) The estimated Peff is used to calculate Fa, which depends on the intestinal transit time and radius [60]:
Fa = 1 - e^(-2 · Peff · T_res / R)T_res= Small intestinal transit time (typically 3 hours or 10,800 seconds)R= Radius of the human small intestine (typically 2 cm)
For a more direct and practical assessment, you can use the following correlation table, which categorizes Papp values into predicted absorption ranges [18]:
Table 1: Correlation between Caco-2 Papp and Predicted Human Intestinal Absorption
| In vitro Papp Value (cm/s) | Predicted Human Fraction Absorbed (Fa) |
|---|---|
| Papp ≤ 1.0 × 10⁻⁶ | Low (0-20%) |
| 1.0 × 10⁻⁶ < Papp ≤ 10 × 10⁻⁶ | Medium (20-70%) |
| Papp > 10 × 10⁻⁶ | High (70-100%) |
FAQ 2: My Caco-2 data shows low recovery. What is the impact and how can I improve it?
Low recovery can significantly impact data interpretation. It may indicate issues like poor solubility, non-specific binding to assay plasticware, cellular metabolism, or compound accumulation within the cell monolayer [40]. This can lead to an underestimation of permeability and mask efflux signals.
To improve recovery:
- Add Bovine Serum Albumin (BSA): Incorporating BSA (e.g., 1.55% w/v) into the assay buffer helps block non-specific binding sites on plasticware and improves the aqueous solubility of lipophilic compounds, leading to more robust permeability and efflux assessment [61] [40].
- Optimize Stirring: Increasing the stirring rate (e.g., to 215 rpm) can reduce the unstirred water layer and improve the permeability of lipophilic compounds [61].
FAQ 3: My laboratory's Caco-2 Papp values for reference compounds differ from literature values. How should I handle this?
Inter-laboratory variability is a recognized challenge in Caco-2 assays [60] [62]. To ensure the reliability of your data and enable accurate cross-study comparisons:
- Use Internal Reference Compounds: Always run a set of high and low permeability reference compounds in parallel with your test compounds. This practice helps normalize for inter-laboratory variability and validates each assay run [18] [60].
- Establish Historical Control Ranges: Maintain a database of Papp values for your reference compounds generated under your specific laboratory conditions. Use this historical data to define acceptable control ranges for your experiments [62].
Table 2: Recommended Reference Compounds for Caco-2 Assay Validation [18]
| Permeability Class | Transporter Role | Example Compound | Typical Test Concentration |
|---|---|---|---|
| Low Permeability | - | Atenolol | 10 µM |
| High Permeability | - | Propranolol/Metoprolol | 10 µM |
| High Permeability | MDR1 (P-gp) Substrate | Digoxin | 10 µM |
| High Permeability | MDR1 (P-gp) Inhibitor | Verapamil | 10 µM |
| High Permeability | BCRP Substrate | Prazosin | 1 µM |
| High Permeability | BCRP Inhibitor | Ko143 | 1 µM |
FAQ 4: How can I predict absorption for highly lipophilic compounds (log P > 3) that often show low Papp in standard assays?
Standard Caco-2 assay conditions can underestimate the permeability of highly lipophilic compounds. Systematic optimization of assay parameters is required [61]. An experimentally optimized design has been shown to improve performance for such compounds:
- BSA Concentration: 1.55% w/v
- Stirring Rate: 215 rpm
- Fed State Simulated Intestinal Fluid: 3.02 mM sodium taurocholate
Using these optimized conditions, the Papp for a compound like octyl paraben (log P 5.69) increased significantly, better reflecting its rapid absorption in humans [61].
Experimental Protocol: Standard Caco-2 Permeability Assay
This protocol provides a detailed methodology for conducting a bidirectional Caco-2 permeability assay to determine Papp and investigate active transport.
Key Materials:
- Cell Model: Ready-to-use Caco-2 monolayers (e.g., CacoReady) or Caco-2 cells (passages 55-70) cultured on Transwell inserts [18] [61].
- Assay Buffer: Hanks' Balanced Salt Solution (HBSS) or similar, with optional 4% w/v BSA for lipophilic compounds [61] [40].
- Control Compounds: Atenolol (low permeability), Propranolol (high permeability), and Digoxin/Talinolol (P-gp substrate) [18] [40].
- Integrity Marker: Lucifer Yellow [40].
- Inhibitors: Verapamil (P-gp inhibitor), Fumitremorgin C or Ko143 (BCRP inhibitor) [18] [40].
Workflow:
Detailed Procedure:
Cell Monolayer Preparation and Integrity Assessment:
- Culture Caco-2 cells on transwell inserts for 21 days to form a confluent, differentiated monolayer. Change the culture medium every two days [18].
- Before the assay, verify monolayer integrity by measuring Transepithelial Electrical Resistance (TEER). Acceptance criteria are typically >1000 Ω·cm² for 24-well plates and >500 Ω·cm² for 96-well plates [18].
- Co-incubate the monolayer with a paracellular marker like Lucifer Yellow. The apparent permeability (Papp) for Lucifer Yellow should be ≤ 1 × 10⁻⁶ cm/s, and the paracellular flux should be ≤ 0.5-0.7% [18] [40].
Test Compound Incubation:
- Prepare the test compound in assay buffer (recommended initial concentration: 10 µM for discovery stages) [18].
- For Apical-to-Basolateral (A-B) transport: Add the compound to the apical donor compartment and collect samples from the basolateral receiver compartment over time (e.g., at 2 hours).
- For Basolateral-to-Apical (B-A) transport: Add the compound to the basolateral donor compartment and collect samples from the apical receiver compartment over time.
- Incubate the system for 2 hours at 37°C with agitation (e.g., 215 rpm) [18] [61].
- Run all compounds in at least triplicate and include reference compounds in each experiment.
Sample Analysis and Data Calculation:
- Analyze the concentration of the test compound in the samples using a sensitive analytical method, such as Liquid Chromatography with tandem Mass Spectrometry (LC-MS/MS) [18] [40].
- Calculate the Apparent Permeability Coefficient (Papp) using the formula:
Papp (cm/s) = (dQ/dt) / (C₀ × A) - Calculate the Efflux Ratio to identify active transport:
Efflux Ratio = Papp (B-A) / Papp (A-B)An efflux ratio > 2 suggests the compound is a substrate for active efflux transporters [40].
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Caco-2 Permeability Assays
| Item | Function / Purpose | Examples / Notes |
|---|---|---|
| Caco-2 Cell Model | In vitro model of the human intestinal epithelium. Forms polarized monolayers with functional transporters. | CacoReady ready-to-use plates; parental Caco-2/ATCC cell line; Caco-2/TC7 clone [18] [62] [9]. |
| Transwell Inserts | Semi-porous filter supports that allow for independent access to apical and basolateral compartments. | Polyester or polycarbonate membranes in 24-well or 96-well formats [18]. |
| Reference Compounds | Validate assay performance and serve as internal controls for permeability and transporter activity. | Atenolol (low passive), Propranolol (high passive), Digoxin (P-gp substrate) [18] [40]. |
| BSA (Bovine Serum Albumin) | Additive to assay buffer to reduce non-specific binding and improve solubility of lipophilic compounds. | Use at 1.5-4% w/v to increase recovery and accuracy for BCS Class II compounds [61] [40]. |
| Transporter Inhibitors | Used to identify specific transporter involvement in compound flux. | Verapamil (P-gp inhibitor); Ko143 (BCRP inhibitor) [18] [40]. |
| LC-MS/MS | Analytical technique for sensitive and specific quantification of compound concentrations in assay samples. | Essential for accurate Papp determination, especially for low-permeability compounds [18] [40]. |
Technical Support Center: Troubleshooting Guides and FAQs
Frequently Asked Questions
Q1: Our Caco-2 models show high variability in permeability measurements between experiments. What could be causing this? A1: Variability in Caco-2 permeability data often stems from several technical factors. Key issues include passage number instability, where higher passage numbers compromise genome stability and alter critical cell characteristics [3]. Additionally, many apparent permeability (Papp) values are dominated by diffusion through unstirred water layers rather than intrinsic membrane permeability [25]. To minimize variability: limit continuous cultures to three months, constantly monitor for changes in phenotype, perform solvent tolerance tests for DMSO concentrations, and ensure subculturing before cells reach 80% confluence to form more homogeneous monolayers [3].
Q2: How can we improve the physiological relevance of our intestinal permeability models beyond standard Caco-2 monolayers? A2: Consider these advanced approaches: Establish co-cultures of Caco-2 and mucin-producing HT29-MTX cells to better replicate the human intestinal environment [9]. Generate "apical-out" organoids that provide direct access to the luminal surface for drug permeability assays [63]. Integrate organoids with microfluidic devices to control flow, gradient formation, and shear stress to better mimic the gut milieu [63]. These approaches address limitations of conventional Caco-2 models, such as the absence of a mucosal layer and oversimplified microenvironment [9].
Q3: What are the major challenges in developing organoid-MPS integrated systems, and how can we address them? A3: The primary challenges include limited survival time due to inadequate vascularization, heterogeneity between organoid batches, and insufficient functional monitoring [64]. Engineering strategies to overcome these limitations include: Using organoid-on-chip technology to precisely control the culture microenvironment through microfluidics [64]. Implementing 3D bioprinting to create more consistent organoid microstructures [65] [66]. Incorporating miniature biochemical sensors to monitor metabolites at micromolar or nanomolar levels with minimal impact on cellular activity [64]. These strategies help bridge the gap between traditional organoid culture and physiologically relevant systems.
Q4: How can we accurately extract intrinsic membrane permeability (P0) from our Caco-2 experiments rather than just apparent permeability (Papp)? A4: Extracting reliable P0 values requires careful experimental design and data analysis. Recent research indicates that only about one quarter of compounds tested in Caco-2/MDCK systems yield reliable P0 values due to various limitations [25]. To improve P0 extraction: Account for possible concentration-shift effects due to different pH values in aqueous layers. Check for limitations posed by aqueous boundary layers, paracellular transport, recovery issues, and active transport processes. Use stricter compound- and reference-specific exclusion criteria during data analysis [25]. Ensure your experimental setup minimizes the impact of unstirred water layers, which dominate most published Papp values.
Troubleshooting Guide for Common Experimental Issues
Issue 1: Slow Growth of Caco-2 Cells
Problem: Caco-2 cells taking too long to grow, significantly dragging out experiments. Solutions:
- Check for mycoplasma contamination, which is a major cause of slow growth.
- Evaluate feeder cell density - an overly dense feeder layer can inhibit growth.
- Subculture cells at 50% confluence rather than 80% to form more homogeneous monolayers [3].
- Consider using accelerated differentiation media or electrospun nanofiber scaffolds to enhance performance [9].
Issue 2: Low TEER Values and Poor Monolayer Integrity
Problem: Inconsistent transepithelial electrical resistance (TEER) measurements indicating compromised barrier function. Solutions:
- For 24-well formats, ensure TEER values >1000 Ω·cm²; for 96-well formats, >500 Ω·cm² [18].
- Verify Lucifer Yellow (LY) apparent permeability ≤ 1 × 10⁻⁶ cm/s and paracellular flux ≤ 0.5-0.7% [18].
- Avoid dome formation and fluid pockets by preventing overcrowding and ensuring even treatment distribution [3].
- Limit passage numbers and culture time to maintain genome stability and critical cell characteristics [3].
Issue 3: High Heterogeneity in Organoid Models
Problem: Significant batch-to-batch variability in organoid formation and function. Solutions:
- Implement automated liquid handling systems for initial stem cell allocation, media addition, and drug testing [64].
- Use synthetic hydrogels or gelatin methacrylate (GelMA) instead of Matrigel to reduce batch variability [67] [64].
- Precisely regulate matrix stiffness and porosity to improve organoid culture outcomes [67].
- Apply engineering tools for precise regulation of medium composition and extracellular matrix innovation [64].
Issue 4: Limited Long-term Viability in MPS
Problem: Inadequate nutrient and oxygen supply affecting long-term growth and functional activities. Solutions:
- Implement oscillating cultures to enhance nutrient access and waste removal [64].
- Incorporate vascularization modules using microfluidic platforms to emulate pulsatile blood luminal flow [65] [66].
- Use droplet-based microfluidic technology with temperature control to generate numerous small organoid spheres from minimal tissue samples [67].
- Consider organoid-on-chip designs that enable close contact between different cell types to capture dynamic cell-cell interplay [65] [66].
Quantitative Data Reference Tables
Table 1: Caco-2 Monolayer Quality Control Standards
| Measurement | CacoReady 24-well Standard | CacoReady 96-well Standard | Acceptance Criteria |
|---|---|---|---|
| TEER | >1000 Ω·cm² | >500 Ω·cm² | Differentiated, polarized cells with formed tight junctions [18] |
| LY Apparent Permeability (Papp) | ≤1×10⁻⁶ cm/s | ≤1×10⁻⁶ cm/s | Intact paracellular barrier [18] |
| LY Paracellular Flux | ≤0.5% | ≤0.7% | Minimal passive paracellular leakage [18] |
Table 2: Predicting In Vivo Absorption from In Vitro Papp Values
| In Vitro Papp Values | Predicted In Vivo Absorption | Interpretation |
|---|---|---|
| Papp ≤ 10⁻⁶ cm/s | Low (0-20%) | Poor intestinal absorption [18] |
| 10⁻⁶ cm/s < Papp ≤ 10×10⁻⁶ cm/s | Medium (20-70%) | Moderate intestinal absorption [18] |
| Papp > 10×10⁻⁶ cm/s | High (70-100%) | Good intestinal absorption [18] |
Table 3: Reference Compounds for Permeability Assay Validation
| Compound Type | Example Compound | Concentration | Purpose |
|---|---|---|---|
| Low Permeability | Atenolol | 10µM | Passive diffusion control [18] |
| High Permeability | Metoprolol | 10µM | Passive diffusion control [18] |
| MDR1 (Pgp) Substrate | Digoxin | 10µM | Transporter activity assessment [18] |
| MDR1 (Pgp) Inhibitor | Verapamil | 10µM | Transporter inhibition control [18] |
| BCRP Substrate | Prazosin | 1µM | Transporter activity assessment [18] |
| BCRP Inhibitor | Ko143 | 1µM | Transporter inhibition control [18] |
Experimental Protocols
Protocol 1: Establishing Patient-Derived Colorectal Organoids for Permeability Studies
Materials:
- Advanced DMEM/F12 medium with antibiotics
- Growth factors: EGF, Noggin, R-spondin1
- Matrigel or synthetic hydrogel alternatives
- Wnt3A and B27 supplement
Method:
- Collect human colorectal tissue samples under sterile conditions immediately following procedures.
- Transfer in cold Advanced DMEM/F12 medium supplemented with antibiotics.
- Process within 6-10 hours or cryopreserve in appropriate medium.
- Isolate crypts through enzymatic digestion and mechanical disruption.
- Embed crypts in Matrigel or synthetic hydrogel and culture with optimized medium.
- For apical-out organoids: apply specific culture conditions to reverse polarity for direct luminal access [63].
- Validate organoids through immunofluorescence staining for cellular characterization.
Critical Steps:
- Prompt tissue processing is essential - delays reduce cell viability and organoid formation efficiency.
- Optimize medium composition to prevent overgrowth of non-tumor cells using specific cytokines like Noggin and B27.
- For co-culture studies, establish immune reconstitution models with autologous immune cells [67].
Protocol 2: Integrating Organoids with Microphysiological Systems
Materials:
- Polydimethylsiloxane (PDMS) for microfluidic device fabrication
- 3D bioprinting equipment (micro-extrusion method)
- Organoid culture in synthetic hydrogels
- Microfluidic pumps for controlled flow rates
Method:
- Fabricate microfluidic devices using soft lithography of PDMS replica or 3D bioprinting.
- Seed organoids in the microfluidic compartments with precise positioning.
- Establish fluid connections between different tissue units for blood supply simulation.
- Apply mechanical stimuli (flow, pressure) to mimic physiological conditions.
- Implement real-time monitoring using optical methods or embedded sensors.
- For permeability studies, create separate compartments representing intestinal lumen and vascular system.
Critical Steps:
- Ensure optical transparency of devices for real-time monitoring of cell migration and interactions.
- Control spatiotemporal gradients of chemicals and mechanical strain within the microfluidic setup.
- Incorporate vascularization modules to emulate pulsatile blood flow, key determinants of tissue functions [65] [66].
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Materials for Organoid-MPS Integration
| Item | Function | Application Notes |
|---|---|---|
| Polydimethylsiloxane (PDMS) | Microfluidic device fabrication | Offers easy fabrication, outstanding optical transparency, and minimal cytotoxicity [65] [66] |
| Synthetic Hydrogels (GelMA) | Extracellular matrix alternative | Provides consistent chemical compositions and physical properties compared to Matrigel [67] |
| Growth Factor Cocktails (Wnt3A, Noggin, R-spondin1) | Stem cell maintenance and differentiation | Promotes growth of various organoids; concentration needs optimization for specific tumor types [63] [67] |
| 3D Bioprinting Systems | Fabrication of organ microstructures | Micro-extrusion is most common method; allows integration of multiple cells, matrix components, and growth factors [65] [66] |
| Miniature Biochemical Sensors | Real-time monitoring of metabolites | Enables monitoring at micromolar or nanomolar levels with minimal impact on cellular activity [64] |
| Automated Liquid Handling Systems | High-throughput organoid culture | Performs precise tasks including stem cell allocation, media changes, and drug testing to reduce heterogeneity [64] |
Workflow and System Architecture Diagrams
Experimental Workflow for Organoid-MPS Integration
Model Evolution: From Caco-2 to Organoid-MPS Platforms
Conclusion
The strategic integration of Matched Molecular Pair Analysis with robust Caco-2 assay data provides a powerful, interpretable framework for optimizing intestinal permeability in drug discovery. This synergistic approach, when combined with emerging machine learning models and advanced in vitro systems like microphysiological platforms, enables researchers to navigate complex structure-permeability relationships with greater confidence. Future advancements will depend on standardizing benchmarking across gut model systems and further refining the integration of computational and experimental data, ultimately closing critical prediction gaps for challenging beyond-Rule-of-Five compounds and accelerating the development of orally bioavailable therapeutics.
References
- 1. Caco-2 - an overview | ScienceDirect Topics [https://www.sciencedirect.com/topics/medicine-and-dentistry/caco-2]
- 2. Should Caco-2 cells retain gold-standard status? | CN Bio [https://cn-bio.com/evaluating-caco-2-cells-gold-standard-status-in-absorption-permeability-and-bioavailability/]
- 3. Caco-2 Cell Tips for Reliable Intestinal Permeability Models [https://mimetas.com/resources/7-ways-caco-2-cells-can-make-or-break-your-intestinal-permeability-models]
- 4. Caco-2 Cell Line - The Impact of Food Bioactives on Health [https://www.ncbi.nlm.nih.gov/books/NBK500149/]
- 5. Caco-2 Cell Growth Essentials: Traits and Troubleshooting ... [https://www.procellsystem.com/resources/cell-culture-academy/caco-2-cell-growth-essentials-traits-and-troubleshooting-tips-1503]
- 6. Re-Use of Caco-2 Monolayers in Permeability Assays ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8537988/]
- 7. Caco-2 cells as a model for intestinal absorption - PubMed [https://pubmed.ncbi.nlm.nih.gov/21400683/]
- 8. Caco-2 Cell Line Standardization with Pharmaceutical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10674574/]
- 9. Advancements in techniques for analyzing cell permeability [https://www.sciencedirect.com/science/article/pii/S2405844025022236]
- 10. ADMET evaluation in drug discovery: 21. Application and ... [https://pubmed.ncbi.nlm.nih.gov/39794857/]
- 11. Semi-automated workflow for molecular pair analysis and ... [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-021-00564-6]
- 12. Matched Molecular Pair Analysis in drug discovery [https://www.sciencedirect.com/science/article/abs/pii/S1359644613000937]
- 13. Using Matched Molecular Series as a Predictive Tool To ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3968889/]
- 14. Matched molecular pair analysis: significance and the ... [https://pubmed.ncbi.nlm.nih.gov/24738976/]
- 15. Matched molecular pair analysis - H1 Connect [https://connect.h1.co/article/718354408]
- 16. ADMET evaluation in drug discovery: 21. Application and industrial validation of machine learning algorithms for Caco-2 permeability prediction | Journal of Cheminformatics | Full Text [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-025-00947-z]
- 17. Matched Molecular Pair Analysis in Short: Algorithms ... [https://www.semanticscholar.org/paper/Matched-Molecular-Pair-Analysis-in-Short%3A-and-Tyrchan-Evertsson/53cc1aba0bf8bfc784dcba6cce1a72c37960540c]
- 18. Caco-2 Permeability Assay Protocol [https://medtechbcn.com/readycell/blog/caco-2-permeability-protocol/]
- 19. How In Vitro DMPK Studies Can Prevent Late-Stage Drug ... [https://blog.crownbio.com/how-in-vitro-dmpk-studies-can-prevent-late-stage-drug-development-failures]
- 20. Optimization of Impedance-Based Real-Time Assay in ... [https://www.mdpi.com/2076-3417/15/15/8298]
- 21. Prediction of blood–brain barrier and Caco-2 permeability ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12051285/]
- 22. Permeation Testing [https://www.teledynelabs.com/products/diffusion/permeation-testing]
- 23. A Case Study from Bayer's Caco-2 Permeability Database [https://pmc.ncbi.nlm.nih.gov/articles/PMC11683871/]
- 24. Improving the Accuracy of Permeability Data to Gain ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11278619/]
- 25. Pitfalls in evaluating permeability experiments with Caco-2/ ... [https://www.sciencedirect.com/science/article/pii/S0928098724000101]
- 26. CaliciBoost: Performance-Driven Evaluation of Molecular ... [https://arxiv.org/html/2506.08059v1]
- 27. Understanding the Caco-2 Permeability Assay [https://www.molecularcloud.org/p/understanding-the-caco-2-permeability-assay-a-critical-tool-in-drug-development]
- 28. ADMET evaluation in drug discovery: 21. Application and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11724520/]
- 29. Assays | ADMET & DMPK | Caco-2 Permeability [https://www.conceptlifesciences.com/assays/caco-2-permeability]
- 30. Improving the Accuracy of Permeability Data to Gain ... [https://www.mdpi.com/2077-0375/14/7/157]
- 31. A Machine Learning Approach for Predicting Caco-2 Cell ... [https://www.mdpi.com/1424-8247/17/6/750]
- 32. QSPR model for Caco-2 cell permeability prediction using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9058322/]
- 33. Categorical Data — xgboost 3.1.1 documentation [https://xgboost.readthedocs.io/en/stable/tutorials/categorical.html]
- 34. How to interpret the output of XGBoost importance? [https://datascience.stackexchange.com/questions/12318/how-to-interpret-the-output-of-xgboost-importance]
- 35. Deployment of XGBoost model to Hugging face space [https://medium.com/@gridflowai/deployment-of-xgboost-model-to-hugging-face-space-50a5c5a8ce59]
- 36. Interpreting XGB feature importance and SHAP values [https://stackoverflow.com/questions/72626523/interpreting-xgb-feature-importance-and-shap-values]
- 37. Steering New Drug Discovery Campaigns: Permeability, Solubility, and Physicochemical Properties in the bRo5 Chemical Space - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7812602/]
- 38. Design Principles for Balancing Lipophilicity and Permeability in beyond Rule of 5 Space - PubMed [https://pubmed.ncbi.nlm.nih.gov/37986275/]
- 39. Matched Molecular Pair Analysis with KNIME [https://www.knime.com/blog/matched-molecular-pair-analysis-with-knime]
- 40. Caco-2 Permeability Assay [https://www.evotec.com/solutions/drug-discovery-preclinical-development/cyprotex-adme-tox-solutions/adme-pk/drug-permeability-and-transporters/caco-2-permeability]
- 41. Conquering the beyond Rule of Five Space with an ... [https://www.mdpi.com/1999-4923/16/7/846]
- 42. a tailored approach to optimize DMPK properties of PROTACs [https://pubs.rsc.org/en/content/articlehtml/2025/md/d4md00854e?page=search]
- 43. Caco-2 Method Validation - Bienta [https://bienta.net/caco-2-assay/caco-2-validation/]
- 44. Effects of Aqueous Boundary Layers and Paracellular ... [https://pubmed.ncbi.nlm.nih.gov/38276501/]
- 45. Paracellular Diffusion in Caco-2 Cell Monolayers: Effect of ... [https://www.sciencedirect.com/science/article/pii/S0022354915503936]
- 46. Unraveling Caco-2 cells through functional and ... [https://www.sciencedirect.com/science/article/abs/pii/S0273230025000017]
- 47. Application of Caco-2 Cell Line in Herb-Drug Interaction ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4355448/]
- 48. Re-Use of Caco-2 Monolayers for a Higher Throughput ... [https://www.preprints.org/manuscript/202405.2057]
- 49. Re-Use of Caco-2 Monolayers in Permeability Assays ... [https://pubmed.ncbi.nlm.nih.gov/34683857/]
- 50. Caco-2 Permeability - Dallas [https://www.utsouthwestern.edu/research/core-facilities/preclinical-pharmacology/pricing-and-services/caco-2-permeability.html]
- 51. First report on machine learning based multiclass ... [https://tandf.figshare.com/articles/dataset/First_report_on_machine_learning_based_multiclass_classification_of_Caco-2_permeability_using_different_balancing_strategies/30073666]
- 52. Handling Imbalanced Data for Classification [https://www.geeksforgeeks.org/machine-learning/handling-imbalanced-data-for-classification/]
- 53. Multiclass Classification With an Imbalanced Data Set [https://builtin.com/machine-learning/multiclass-classification]
- 54. Methods for multi‑class imbalanced data classification [https://medium.com/@Jerrylzj/methods-for-multi-class-imbalanced-data-classification-574ab4b73d09]
- 55. Reliable Prediction of Caco-2 Permeability by Supervised ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9610902/]
- 56. Machine learning small molecule properties in drug ... [https://www.sciencedirect.com/science/article/pii/S2949747723000209]
- 57. Benchmarking community drug response prediction models [https://arxiv.org/html/2503.14356v1]
- 58. Predicting Caco-2/MDCK Intrinsic Membrane Permeability ... [https://www.sciencedirect.com/science/article/pii/S0928098725002787]
- 59. MembranePlus™ - Simulations Plus Caco-2 Cells | In Vitro ... [https://www.simulations-plus.com/software/membraneplus/]
- 60. Predicting human pharmacokinetics from preclinical data [https://pmc.ncbi.nlm.nih.gov/articles/PMC7533162/]
- 61. Optimisation of the caco-2 permeability assay using ... [https://pubmed.ncbi.nlm.nih.gov/18293061/]
- 62. An exploratory study of two Caco-2 cell models for oral ... [https://pubmed.ncbi.nlm.nih.gov/21105755/]
- 63. A Practical Guide to Developing and Troubleshooting ... [https://www.mdpi.com/2409-9279/8/5/121]
- 64. Strategies to overcome the limitations of current organoid technology - engineered organoids - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12033597/]
- 65. Frontiers | Organoids and Microphysiological Systems: New Tools for Ophthalmic Drug Discovery [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2020.00407/full]
- 66. Organoids and Microphysiological Systems: New Tools for Ophthalmic Drug Discovery - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC7147294/]
- 67. Breakthroughs and challenges of organoid models for ... [https://www.nature.com/articles/s41420-025-02505-w]