Bridging the Prediction Gap: A Practical Guide to Validating In Silico ADMET Models with In Vitro Data
This article provides a comprehensive framework for researchers and drug development professionals to effectively validate in silico ADMET predictions with robust in vitro data.
Bridging the Prediction Gap: A Practical Guide to Validating In Silico ADMET Models with In Vitro Data
Abstract
This article provides a comprehensive framework for researchers and drug development professionals to effectively validate in silico ADMET predictions with robust in vitro data. As the pharmaceutical industry increasingly relies on computational tools to accelerate discovery, bridging the gap between prediction and experimental validation is critical for reducing late-stage attrition. We explore the foundational principles of ADMET modeling, detail advanced methodological and integrated application workflows, address common troubleshooting and optimization challenges, and present rigorous validation and comparative strategies. By synthesizing current trends, including the use of AI and complex in vitro models, this guide aims to enhance the reliability and regulatory acceptance of in silico-in vitro integrated approaches in preclinical development.
The Critical Need for ADMET Validation in Modern Drug Discovery
Why Predictive ADMET is a Cornerstone for Reducing Clinical Attrition
The pharmaceutical industry faces a formidable challenge: the overwhelming majority of drug candidates fail to reach the market, often after substantial investments have been made in clinical trials. A primary reason for these late-stage failures is unsatisfactory profiles in Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET). Poor ADMET properties account for approximately 50-60% of clinical phase failures, highlighting a critical bottleneck in the drug development pipeline [1] [2]. This high attrition rate translates into staggering costs, with the average new drug requiring nearly $2.6 billion and 12-15 years to develop [3].
The traditional drug discovery approach, which tested ADMET properties relatively late in the process, has proven economically and temporally unsustainable. Consequently, the industry has undergone a significant strategic shift, now performing extensive ADMET screening considerably earlier in the drug discovery process [1]. This paradigm shift positions predictive ADMET as a cornerstone strategy for de-risking drug development. By identifying and eliminating problematic compounds before they enter costly clinical phases, in silico (computational) ADMET prediction saves both money and time, boosting overall medication development efficiency [1]. For researchers and drug development professionals, this translates into a workflow where computational models are no longer auxiliary tools but fundamental components for making critical go/no-go decisions on candidate compounds.
The Evolution and Methodologies ofIn SilicoADMET Prediction
The field of in silico ADMET prediction has evolved dramatically from simple quantitative structure-activity relationship (QSAR) models to sophisticated artificial intelligence (AI) and machine learning (ML) platforms. This evolution has been driven by the accumulation of large-scale experimental data and advances in computational algorithms.
Fundamental Computational Approaches
At its core, predictive ADMET relies on a suite of computational methods that span different levels of complexity and mechanistic insight:
- Quantum Mechanics (QM) and Molecular Mechanics (MM): These methods provide deep mechanistic insights, particularly for understanding metabolic processes. For instance, QM/MM simulations have been used to study the metabolism of camphor by bacterial P450 enzymes and to examine the regioselectivity of estrone metabolism in humans, helping to predict which specific positions in a molecule are most susceptible to oxidation by CYP enzymes [1].
- Quantitative Structure-Activity Relationship (QSAR/QSPR): These traditional yet powerful approaches establish quantitative relationships between molecular descriptors (physicochemical properties, topological structures, etc.) and ADMET endpoints. Commercial tools like ADMET Predictor extensively use QSPR models, constructing predictions for over 220 properties that comprehensively cover a compound's absorption, distribution, metabolism, excretion, toxicity, and pharmacokinetic parameters [4].
- Molecular Docking and Pharmacophore Modeling: These techniques are particularly valuable for predicting specific interactions, such as a compound's binding to metabolic enzymes (e.g., CYP450) or toxicity-related receptors like the hERG channel [1].
The Rise of Machine Learning and AI
Modern ADMET prediction has been revolutionized by machine learning and artificial intelligence, which can model complex, non-linear structure-property relationships that are difficult to capture with traditional methods [5]. Key ML approaches include:
- Graph Neural Networks (GNNs): These methods represent molecules as graphs (atoms as nodes, bonds as edges) and can directly learn from molecular structure without relying on pre-defined descriptors. Platforms like ADMETlab 3.0 use Directed Message Passing Neural Networks (DMPNN) to achieve state-of-the-art prediction performance [6].
- Multitask Learning (MTL): This framework allows simultaneous prediction of multiple ADMET endpoints by sharing representations across related tasks, improving data efficiency and model robustness [5].
- Ensemble Methods: These combine predictions from multiple base models (e.g., decision trees, neural networks) to create more robust and accurate consensus predictions, effectively addressing challenges like limited data availability and high model uncertainty [5].
The performance of these models is heavily dependent on the quality and quantity of training data. Leading platforms are trained on massive datasets; for example, ADMETlab 3.0 incorporates over 400,000 data entries across 119 endpoints, while some commercial tools additionally integrate proprietary data from pharmaceutical industry partners to enhance model accuracy [4] [6].
Comparative Analysis of Leading ADMET Prediction Platforms
The landscape of ADMET prediction tools is diverse, ranging from open-source packages to comprehensive commercial suites. The table below provides a structured comparison of several prominent platforms, highlighting their respective capabilities, data coverage, and primary applications.
Table 1: Comparison of Leading ADMET Prediction Platforms
| Platform Name | Type/Availability | Key Features | Model Foundation | Endpoint Coverage | Primary Applications |
|---|---|---|---|---|---|
| ADMET Predictor [4] | Commercial Software | - ADMET Risk scoring based on 2,260 marketed drugs- High-throughput PK (HTPK) module- Structure Sensitivity Analysis (SSA)- Predicts 220+ properties | QSPR models combined with AI algorithms | Covers physicochemical properties, transporters, metabolism, toxicity, and PK parameters | Early compound screening, formulation design, toxicity mitigation, dose prediction |
| ADMETlab 3.0 [6] | Free Web Platform | - Covers 119 endpoints (77 modeled, 42 calculated)- API for programmatic access- Uncertainty estimation for predictions- Molecular comparison tools | DMPNN-Des (Graph Neural Network + RDKit 2D descriptors) | Extensive coverage of physicochemical, ADME, and toxicity properties | Academic research, virtual screening, lead optimization |
| ADMET-AI [7] | Open-Source | - Fast prediction speed- Benchmarks against approved drugs in DrugBank- Provides percentile ranks relative to reference drugs- Easy integration via Python API | Chemprop-RDKit Graph Neural Network | 41 ADMET datasets from TDC (Therapeutics Data Commons) | Early-stage compound prioritization, relative risk assessment |
| SwissADME [6] | Free Web Tool | - User-friendly interface- Key physicochemical and pharmacokinetic descriptors- Drug-likeness rules (e.g., Lipinski, Veber) | Combination of rule-based and ML models | Focuses on key physicochemical properties and absorption-related parameters | Quick initial profiling, educational use |
Performance and Accuracy Considerations
When evaluating these platforms, predictive accuracy remains the paramount criterion. Independent literature validations have shown that several commercial and academic tools achieve high performance. For instance, ADMET Predictor's models for key properties like logP (a measure of lipophilicity), fraction unbound in plasma (fup), and P-gp substrate identification have demonstrated strong concordance with experimental data [4]. Similarly, ADMETlab 3.0 reports R² values for regression tasks primarily between 0.75 and 0.95, and AUC values for classification tasks ranging from 0.72 to 0.99, indicating robust predictive power across diverse endpoints [6].
A critical differentiator among modern platforms is the inclusion of Uncertainty Quantification (UQ). Tools like ADMETlab 3.0 and ADMET-AI provide estimates of prediction confidence, which is crucial for prioritizing compounds in virtual screening. ADMETlab 3.0 implements an evidence-based approach for regression models and Monte Carlo dropout for classification models to assess uncertainty [6]. This functionality helps researchers identify when a prediction is outside the model's reliable "applicability domain," reducing the risk of decisions based on unreliable forecasts.
ValidatingIn SilicoPredictions withIn VitroandIn VivoData
The true test of any in silico model is its ability to correlate with empirical data. The validation of ADMET predictions follows a hierarchical approach, moving from in vitro assays to in vivo studies, with each step providing a more complex layer of confirmation.
Key Experimental Protocols for Validation
To establish a robust validation framework for in silico ADMET predictions, researchers employ a suite of standardized experimental protocols. The table below details key methodologies that serve as benchmarks for computational forecasts.
Table 2: Key Experimental Protocols for Validating ADMET Predictions
| ADMET Property | Experimental Protocol | Brief Description & Function | Key Output Metrics |
|---|---|---|---|
| Absorption | Caco-2 Permeability Assay [8] | Uses human colon adenocarcinoma cell monolayers to model intestinal absorption. | Apparent Permeability (Papp), predicts absorption rate (ka) and fraction absorbed (Fa). |
| Metabolism | Liver Microsome/Hepatocyte Stability [8] | Incubates test compound with liver enzymes to measure metabolic degradation. | Intrinsic Clearance (CLint), used in IVIVE to predict in vivo hepatic clearance (CL). |
| Toxicity | hERG Inhibition Assay [7] | Measures compound's potential to block the hERG potassium channel, linked to cardiac arrhythmia. | IC50 value (concentration causing 50% inhibition); predictive of Torsades de Pointes risk. |
| Distribution | Plasma Protein Binding [8] | Determines the fraction of drug bound to plasma proteins vs. free (pharmacologically active). | Fraction unbound (fup); critical for correcting clearance and volume of distribution predictions. |
| Distribution | P-gp Transporter Assay [4] | Evaluates if a compound is a substrate or inhibitor of the P-glycoprotein efflux transporter. | Efflux ratio; predicts potential for drug-drug interactions and tissue penetration (e.g., BBB). |
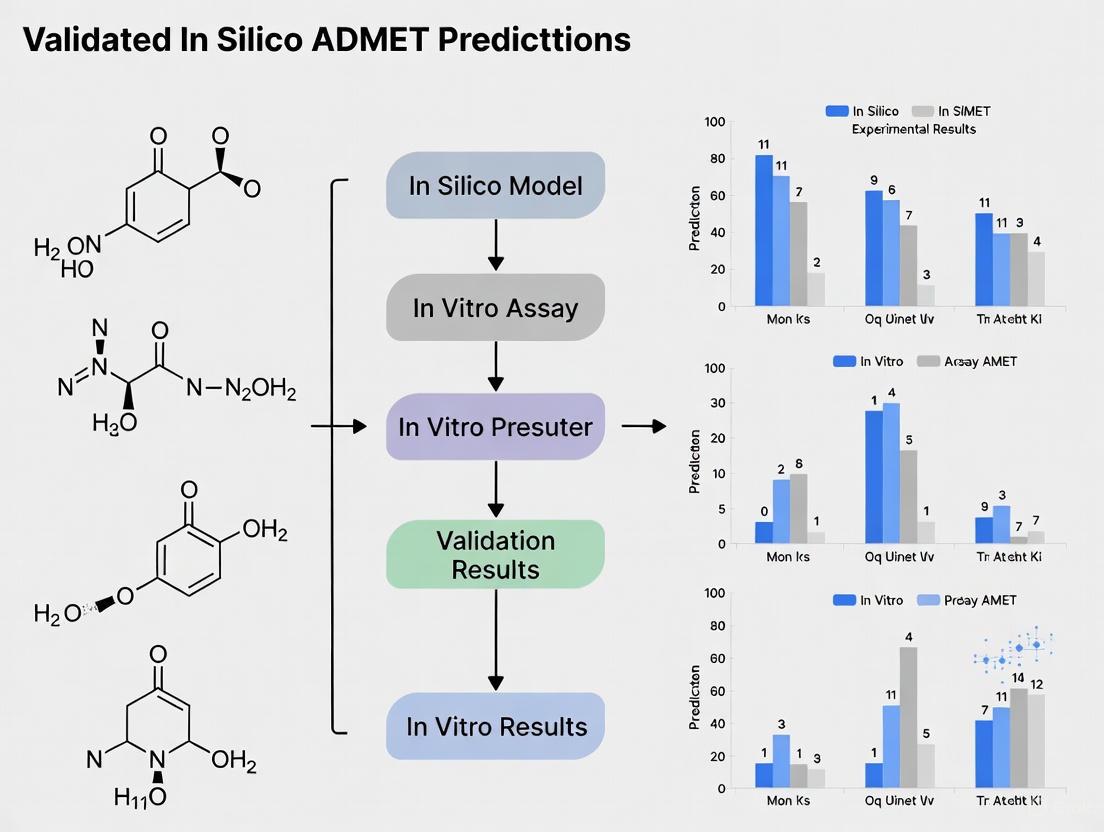
These experimental protocols provide the essential ground-truth data against which in silico predictions are validated. The relationship between computational prediction and experimental validation can be visualized as an iterative cycle that refines model accuracy and informs drug design.
Diagram Title: ADMET Prediction and Validation Workflow
Case Studies: Integrating Prediction with Experimental Data
Concrete examples illustrate the power and limitations of integrating in silico predictions with experimental data:
GSK3β Inhibitors and hERG Toxicity: A study using the open-source tool ADMET-AI demonstrated its ability to predict the hERG cardiotoxicity risk of two GSK3β inhibitors. The tool correctly predicted a high probability (0.98) of hERG inhibition for a problematic compound (Cmpd 1, experimental hERG IC50 = 44 nM) and a lower probability (0.73) for an optimized analog (Cmpd 14). However, it is noteworthy that Cmpd 14 was still classified as a hERG inhibitor by the model despite an experimental IC50 >100 µM, highlighting a potential area for model refinement concerning negative prediction accuracy [7].
MET Inhibitor and CYP3A4 Time-Dependent Inhibition (TDI): ADMET-AI was used to retrospectively predict the CYP3A4 inhibition risk for a MET inhibitor (compound 13) and its N-desmethyl metabolite. The model predicted a high probability of CYP3A4 inhibition for both, especially the metabolite (0.849), corroborating experimental findings that the metabolite was a potent inhibitor (Ki = 105 nM). This case shows how in silico tools can predict metabolic activation leading to toxicity, a critical consideration in drug design [7].
These case studies underscore that while in silico tools are powerful for risk stratification and prioritization, they are most effective when used in concert with experimental data rather than as standalone arbiters.
The Scientist's Toolkit: Essential Research Reagents and Materials
Validating in silico ADMET predictions requires a well-characterized set of biological reagents and assay systems. The following table details key materials essential for conducting the experimental protocols outlined in the previous section.
Table 3: Essential Research Reagent Solutions for ADMET Experimental Validation
| Research Reagent / Material | Function in ADMET Assessment |
|---|---|
| Caco-2 Cell Line [8] | A model of the human intestinal epithelium used to predict oral absorption and permeability of drug candidates. |
| Human Liver Microsomes (HLM) / Cryopreserved Hepatocytes [8] | Enzyme systems derived from human liver tissue used to study metabolic stability, clearance, and metabolite identification. |
| HEK293 Cells Expressing hERG Channel [7] | A cell line engineered to express the human Ether-à-go-go Related Gene potassium channel, crucial for assessing cardiotoxicity risk. |
| Human Plasma [8] | Used in equilibrium dialysis or ultrafiltration experiments to determine the extent of plasma protein binding (fraction unbound, fup). |
| MDR1-MDCK II Cell Line [4] | Canine kidney cells expressing the human P-glycoprotein (MDR1) transporter, used to assess efflux potential and blood-brain barrier penetration. |
Predictive ADMET has unequivocally established itself as a cornerstone for reducing clinical attrition. By enabling the early identification of compounds with unfavorable pharmacokinetic and safety profiles, in silico tools directly address the leading cause of failure in drug development. The continuous improvement of AI and ML models, coupled with the expansion of high-quality biological data, is steadily increasing the accuracy and reliability of these predictions. The future of the field lies in the tighter integration of computation and experimentation, where in silico predictions not only guide experimental design but are also continuously refined by experimental results. This virtuous cycle, supported by robust validation protocols and a clear understanding of each tool's strengths and limitations, promises to streamline the drug development pipeline, increase success rates, and ultimately accelerate the delivery of safer and more effective medicines to patients.
The assessment of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties represents a critical gatekeeper in drug discovery, determining whether promising drug candidates succeed or fail during development. Approximately 40-45% of clinical attrition continues to be attributed to ADMET liabilities, making accurate prediction of these properties essential for improving drug development efficiency [9]. The pharmaceutical industry employs three complementary methodological approaches—in silico, in vitro, and in vivo—each with distinct advantages, limitations, and applications within the drug development pipeline. These approaches form an interconnected toolkit that enables researchers to evaluate how compounds behave within biological systems, from initial screening through preclinical development.
Over decades, ADMET properties have become one of the most important issues for assessing the effects or risks of small molecular compounds on the human body [10]. The growing need to minimize animal use in medical development and research further highlights the increasing significance of in silico and in vitro tools [1]. This guide provides a comprehensive comparison of these three methodological landscapes, focusing on their respective roles, experimental protocols, and how their integration—particularly the validation of in silico predictions with in vitro data—strengthens the drug discovery process.
Core Methodologies: Definitions and Characteristics
Comparative Analysis of ADMET Approaches
Table 1: Fundamental characteristics of in silico, in vitro, and in vivo ADMET evaluation methods
| Feature | In Silico | In Vitro | In Vivo |
|---|---|---|---|
| Definition | Computational simulation of ADMET properties | Experiments conducted in controlled laboratory environments using biological components outside living organisms | Studies performed within living organisms |
| Throughput | Very high (can screen thousands of compounds rapidly) | Moderate to high (depends on assay format) | Low (time-intensive and resource-heavy) |
| Cost Factors | Very low once models are established | Moderate (reagents, equipment, labor) | Very high (animal costs, facilities, personnel) |
| Time Requirements | Minutes to hours for predictions | Days to weeks depending on assay complexity | Weeks to months for complete studies |
| Data Output | Predictive parameters and calculated properties | Quantitative measurements of specific processes | Integrated physiological responses |
| Regulatory Acceptance | Supporting role for decision-making | Accepted for specific endpoints (e.g., Caco-2 for permeability) | Gold standard for preclinical safety |
| Key Advantages | No physical samples required; high speed and low cost [1] | Controlled environment; mechanistic insights | Complete physiological context |
| Primary Limitations | Model dependency and applicability domain constraints [10] | Simplified biological representation | Species translation challenges; ethical considerations |
Methodology-Specific Applications
In silico approaches eliminate the need for physical samples and laboratory facilities while providing rapid and cost-effective alternatives to expensive and time-consuming experimental testing [1]. These computational methods include quantum mechanics calculations, molecular docking, pharmacophore modeling, QSAR analysis, molecular dynamics simulations, and PBPK modeling [1]. The fusion of Artificial Intelligence (AI) with computational chemistry has further revolutionized drug discovery by enhancing compound optimization, predictive analytics, and molecular modeling [11].
In vitro models include systems such as Caco-2 cell monolayers for intestinal permeability assessment, which have emerged as the "gold standard" for drug permeability due to their ability to closely mimic the human intestinal epithelium [12]. These assays provide a balance between biological relevance and experimental control, though they may not fully capture the complexity of whole organisms.
In vivo studies remain essential for understanding complete pharmacokinetic profiles and toxicity outcomes in intact physiological systems. However, there is growing pressure to reduce animal testing through the principles of the 3Rs (Replacement, Reduction, and Refinement), driving increased adoption of in silico and in vitro alternatives [1].
Experimental Validation: Integrating In Silico Predictions with In Vitro Data
Validation Workflow and Relationship
The convergence of in silico predictions with experimental validation represents a cornerstone of modern ADMET evaluation. The following diagram illustrates the systematic workflow for validating computational predictions with biological assays:
Diagram 1: Integrated ADMET validation workflow showing the feedback loop between methodologies
This validation framework creates a virtuous cycle where computational models identify promising candidates for experimental testing, and experimental results subsequently refine and improve the computational models. The feedback loop is essential for enhancing model accuracy and expanding applicability domains over time.
Case Study: Caco-2 Permeability Prediction
The Caco-2 cell model has been widely used to assess intestinal permeability of drug candidates in vitro, owing to its morphological and functional similarity to human enterocytes [12]. This validation case study exemplifies the rigorous comparison between computational predictions and experimental measurements.
Table 2: Performance comparison of machine learning models for Caco-2 permeability prediction
| Model Type | Dataset Size | Key Features | Performance Metrics | Reference |
|---|---|---|---|---|
| XGBoost | 5,654 compounds | Morgan fingerprints + RDKit2D descriptors | Best overall performance on test sets | [12] |
| Random Forest | 5,654 compounds | Morgan fingerprints + RDKit2D descriptors | Competitive performance | [12] |
| Message Passing Neural Network (MPNN) | 5,654 compounds | Molecular graph representation | Captured nuanced molecular features | [12] |
| Boosting Model | 1,272 compounds | MOE 2D/3D descriptors | R² = 0.81, RMSE = 0.31 | [12] |
| MESN Deep Learning | 4,464 compounds | Multiple molecular embeddings | MAE = 0.410, RMSE = 0.545 | [12] |
| Consensus Random Forest | 4,900+ molecules | QSPR approach with feature selection | RMSE = 0.43-0.51, R² = 0.57-0.61 | [12] |
Experimental Protocol for Caco-2 Validation:
- Data Curation: Compile experimental Caco-2 permeability measurements from public sources and in-house datasets
- Molecular Standardization: Apply standardized procedures for tautomer canonicalization and neutral forms preservation
- Representation Generation: Calculate multiple molecular representations including Morgan fingerprints, RDKit 2D descriptors, and molecular graphs
- Model Training: Implement various machine learning algorithms with scaffold-based data splitting to ensure structural diversity
- Validation: Assess model performance using hold-out test sets and external validation compounds from industrial collections
- Application Domain Analysis: Evaluate model robustness and define boundaries for reliable predictions [12]
This systematic validation approach demonstrates that machine learning models, particularly XGBoost, can achieve significant predictive accuracy for Caco-2 permeability, enabling their use as reliable tools for assessing intestinal absorption during early-stage drug discovery [12].
Advanced Approaches and Recent Innovations
Federated Learning for Expanded Chemical Space Coverage
A fundamental challenge in ADMET prediction is that model performance typically degrades when predictions are made for novel scaffolds or compounds outside the distribution of training data [9]. Federated learning addresses this limitation by enabling model training across distributed proprietary datasets without centralizing sensitive data, thus expanding the chemical space coverage.
Cross-pharma research has demonstrated that federated models systematically outperform local baselines, and performance improvements scale with the number and diversity of participants [9]. This approach alters the geometry of chemical space a model can learn from, improving coverage and reducing discontinuities in the learned representation. The benefits persist across heterogeneous data, as all contributors receive superior models even when assay protocols, compound libraries, or endpoint coverage differ substantially [9].
Quantitative In Vitro to In Vivo Extrapolation (QIVIVE)
Quantitative in vitro to in vivo extrapolation has emerged as a crucial methodology for converting concentrations that produce adverse outcomes in vitro to corresponding in vivo doses using physiologically based kinetic modeling-based reverse dosimetry [13]. A significant challenge in applying QIVIVE arises from the common use of "nominal" chemical concentrations reported for in vitro assays that are not directly comparable to "free" chemical concentrations in plasma observed in vivo [13].
Recent comparative analyses of chemical distribution models have evaluated the performance of different in vitro mass balance models for predicting free media or cellular concentrations [13]. These studies found that predictions of media concentrations were more accurate than those for cells, and that the Armitage model had slightly better performance overall [13]. Through sensitivity analyses, researchers determined that chemical property-related parameters were most influential for media predictions, while cell-related parameters were also important for cellular predictions.
AI and Machine Learning Advancements
The integration of Artificial Intelligence (AI) with traditional computational methods has transformed ADMET prediction landscapes. Core AI algorithms including support vector machines, random forests, graph neural networks, and transformers are now extensively applied in molecular representation, virtual screening, and ADMET property prediction [11]. Platforms like Deep-PK and DeepTox leverage graph-based descriptors and multitask learning for pharmacokinetics and toxicity prediction [11].
In structure-based design, AI-enhanced scoring functions and binding affinity models outperform classical approaches, while deep learning transforms molecular dynamics by approximating force fields and capturing conformational dynamics [11]. The convergence of AI with quantum chemistry and density functional theory is illustrated through surrogate modeling and reaction mechanism prediction, though challenges remain in data quality, model interpretability, and generalizability [11].
Essential Research Tools and Reagents
Research Reagent Solutions for ADMET Evaluation
Table 3: Key research reagents and computational tools for ADMET assessment
| Tool/Category | Specific Examples | Primary Function | Application Context |
|---|---|---|---|
| Cell-Based Assay Systems | Caco-2, MDCK, LLC-PK1 cell lines | Model intestinal, renal, and blood-brain barrier permeability | In vitro permeability assessment [12] |
| Computational Chemistry Software | Quantum mechanics (QM), Molecular mechanics (MM) | Predict reactivity, stability, and metabolic routes | In silico ADMET profiling [1] |
| Molecular Representations | Morgan fingerprints, RDKit 2D descriptors, molecular graphs | Encode structural features for machine learning | Model training and prediction [12] [14] |
| Machine Learning Algorithms | XGBoost, Random Forest, MPNN, SVM | Build predictive models from structural data | In silico property prediction [12] [14] |
| Metabolic Enzyme Systems | CYP3A4, CYP2D6, CYP2C9, CYP2C19 | Assess compound metabolism and potential interactions | In vitro and in silico metabolism studies [1] |
| Physiologically-Based Kinetic Models | PBK modeling with reverse dosimetry | Convert in vitro concentrations to in vivo doses | QIVIVE implementation [13] |
| In Vitro Mass Balance Models | Armitage, Fischer, Fisher models | Predict free concentrations in assay media | In vitro assay interpretation [13] |
The integration of in silico, in vitro, and in vivo methodologies represents the most promising path forward for comprehensive ADMET evaluation in drug discovery. While each approach has distinct strengths and limitations, their synergistic application creates a powerful framework for predicting compound behavior and mitigating late-stage attrition. The continuous refinement of computational models through experimental validation, as demonstrated in the Caco-2 permeability case study, enables increasingly accurate predictions that can guide compound selection and optimization during early discovery phases.
Recent advances in federated learning, AI-powered predictive modeling, and quantitative in vitro to in vivo extrapolation are addressing fundamental challenges in chemical space coverage, data diversity, and physiological relevance. As these methodologies continue to evolve and integrate, the drug discovery community moves closer to developing truly generalizable ADMET models with expanded predictive power across the chemical and biological diversity encountered in modern pharmaceutical research. This progression ultimately supports the development of safer, more effective therapeutics while potentially reducing costs and animal testing in the drug development pipeline.
Table of Contents
- Introduction: The ADMET Failure Landscape
- Quantifying the Impact: ADMET Attrition in the Drug Development Pipeline
- Benchmarking Predictive Models: Performance on Key ADMET Properties
- A Guide to Experimental Protocols for ADMET Validation
- The Scientist's Toolkit: Essential Reagents and Resources
- Integrated Workflows: Bridging In Silico and In Vitro Data
- Future Perspectives in ADMET Prediction
Drug discovery and development is a high-stakes endeavor, plagued by considerable doubt and a high likelihood of failure. A leading cause of this failure is undesirable Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties [15] [16]. Understanding the pharmacokinetics, safety, and efficacy of candidate drugs is crucial for their success, requiring early assessment of these properties in the discovery process [15]. Poor ADMET profiles are a major cause of attrition in drug development, accounting for approximately 40% of compound failures during the testing phase [17]. This translates to a massive financial burden, with the average cost of developing a single new drug estimated to easily exceed $2 billion [15]. The high attrition rate underscores the critical need for robust tools to predict and validate ADMET properties early, weeding out problematic compounds before they enter the costly clinical development phase [16].
Quantifying the Impact: ADMET Attrition in the Drug Development Pipeline
The following table summarizes the key ADMET parameters and their role in drug failure, highlighting why they are critical to assess early.
Table 1: Key ADMET Properties and Their Impact on Drug Failure
| ADMET Property | Description | Consequence of Poor Performance | Common Experimental Models |
|---|---|---|---|
| Absorption | Transportation of the unmetabolized drug from the administration site to circulation [18]. | Low oral bioavailability, inadequate therapeutic effect [18]. | Caco-2 cell permeability, Human Intestinal Absorption (HIA) models [19] [18]. |
| Distribution | Reversible transfer of a drug through the body's blood and tissues [18]. | Failure to reach the target site of action (e.g., brain), or distribution to sensitive tissues causing toxicity [18]. | Blood-Brain Barrier (BBB) penetration, Plasma Protein Binding (PPB) [18]. |
| Metabolism | Biotransformation of the drug in the body [18]. | Too rapid metabolism leads to short duration of action; too slow metabolism leads to accumulation and toxicity [20]. | Cytochrome P450 (CYP) inhibition/induction, Human Liver Microsomal (HLM) stability [20] [18]. |
| Excretion | The removal of the administered drug from the body [18]. | Accumulation of the drug, leading to potential toxicity [18]. | Clearance (Cl), Half-life (t1/2) [18]. |
| Toxicity | The level of damage a compound can inflict on an organism [18]. | Adverse effects in patients, drug withdrawal from the market, trial failure [16]. | hERG inhibition (cardiac toxicity), Ames test (mutagenicity), carcinogenicity [18]. |
Benchmarking Predictive Models: Performance on Key ADMET Properties
The field of in silico ADMET prediction has evolved significantly, with various machine learning (ML) and deep learning (DL) models now offering rapid, cost-effective screening. The table below compares the performance of different modeling approaches on benchmark ADMET tasks, using standard evaluation metrics.
Table 2: Performance Comparison of In Silico ADMET Prediction Models
| Prediction Task | Model Type | Dataset | Key Performance Metrics | Citation |
|---|---|---|---|---|
| Caco-2 Permeability | XGBoost (on Morgan Fingerprints + RDKit 2D Descriptors) | 5,654 compounds | Best-performing model on test sets vs. RF, GBM, SVM, DMPNN, and CombinedNet. | [19] |
| Caco-2 Permeability | Message Passing Neural Network (MPNN) | 4,464 compounds | MAE = 0.410, RMSE = 0.545 | [19] |
| CYP450 Inhibition | Attention-based Graph Neural Network (GNN) | Six benchmark datasets | Competitive performance on CYP2C9, CYP2C19, CYP2D6, and CYP3A4 classification tasks. | [15] |
| Aqueous Solubility (log S) | Attention-based Graph Neural Network (GNN) | Six benchmark datasets | Effective performance on regression task, bypassing molecular descriptors. | [15] |
| Lipophilicity (log P) | Attention-based Graph Neural Network (GNN) | Six benchmark datasets | Effective performance on regression task, bypassing molecular descriptors. | [15] |
| Multi-task ADME-T | Transformer-based Model | Pre-trained on 1.8B molecules from ZINC/PubChem | High accuracy in predicting a wide array of properties (e.g., solubility, BBB penetration, toxicity). | [17] |
Evaluation Metrics Explained:
- Classification Models: Accuracy, Precision, Recall, F1-score (range 0-1, higher is better), and ROC-AUC (area under the receiver operating characteristic curve) are used to evaluate models that categorize compounds (e.g., CYP inhibitor vs. non-inhibitor) [20].
- Regression Models: Mean Absolute Error (MAE), Root Mean Squared Error (RMSE) (lower is better), and the Coefficient of Determination (R²) (higher is better, up to 1) are used for models predicting continuous values (e.g., solubility, permeability values) [19] [20].
A Guide to Experimental Protocols for ADMET Validation
For a computational prediction to be trusted, it must be validated with experimental data. Below are detailed methodologies for key assays cited in the literature.
Caco-2 Permeability Assay for Predicting Intestinal Absorption
The Caco-2 cell model is the "gold standard" for assessing intestinal permeability in vitro due to its morphological and functional similarity to human enterocytes [19].
Protocol:
- Cell Culture: Culture Caco-2 cells in standard media (e.g., DMEM with 10% FBS, 1% non-essential amino acids, and 1% penicillin-streptomycin) [19].
- Seeding and Differentiation: Seed cells on permeable filter supports in transwell plates. Allow cells to differentiate and form tight junctions over 21 days [19].
- Transepithelial Transport: On the day of the experiment, add the test compound to the donor compartment (apical side for absorption study). The receiver compartment (basolateral side) contains fresh buffer [19].
- Sampling and Analysis: Incubate for a set time (e.g., 2 hours). Sample from the receiver compartment and analyze compound concentration using a sensitive method like Liquid Chromatography-Mass Spectrometry (LC-MS) or High-Performance Liquid Chromatography (HPLC) [19].
- Data Calculation: Calculate the apparent permeability coefficient (Papp) using the formula: Papp = (dQ/dt) / (A × C0), where dQ/dt is the transport rate, A is the membrane surface area, and C0 is the initial donor concentration [19]. Results are often reported as log Papp (cm/s).
Molecular Docking for Binding Affinity and Metabolism Studies
Molecular docking is a computational method used to predict the orientation and binding affinity of a small molecule (ligand) within a protein's active site, useful for understanding interactions with metabolic enzymes like CYP450s [21].
Protocol:
- Protein Preparation: Obtain the 3D structure of the target protein (e.g., from Protein Data Bank, PDB). Remove water molecules and co-crystallized ligands. Add hydrogen atoms and assign partial charges using software like MOE or Schrödinger Suite [21].
- Ligand Preparation: Draw or obtain the 3D structure of the test molecule. Energy-minimize the structure and generate potential 3D conformations [21].
- Docking Simulation: Define the active site (often based on the location of a co-crystallized native ligand). Run the docking algorithm to generate multiple binding poses. The software scores each pose based on an energy function [21].
- Validation and Analysis: Validate the docking procedure by re-docking the native ligand and calculating the Root-Mean-Square Deviation (RMSD) between the predicted and original pose; an RMSD < 2.0 Å is acceptable. Analyze the binding mode (poses) and interactions (hydrogen bonds, hydrophobic interactions) of the test compound [21].
MTT Cytotoxicity Assay
The MTT assay is a colorimetric method for assessing cell metabolic activity, used as a proxy for cell viability and compound toxicity [21].
Protocol:
- Cell Seeding: Seed adherent cells (e.g., human gingival fibroblasts) in a 96-well plate and allow them to attach overnight [21].
- Compound Treatment: Treat cells with a range of concentrations of the test compound. Include negative control (vehicle only) and positive control (a known toxic compound) wells [21].
- MTT Incubation: After a designated exposure time (e.g., 24-72 hours), add MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) to each well and incubate for 2-4 hours. Living cells reduce the yellow MTT to purple formazan crystals [21].
- Solubilization and Measurement: Remove the media and dissolve the formazan crystals in a solvent like DMSO. Measure the absorbance of the solution at 570 nm using a microplate reader [21].
- Data Analysis: Calculate the percentage of cell viability compared to the negative control. The concentration that inhibits cell growth by 50% (IC50) is determined from the dose-response curve [21].
Successful ADMET validation relies on a suite of computational and experimental resources. The following table details key tools for modern drug discovery research.
Table 3: Research Reagent Solutions for ADMET Validation
| Item / Resource | Function / Description | Example Use in ADMET Validation |
|---|---|---|
| Caco-2 Cell Line | A human colon adenocarcinoma cell line that differentiates into enterocyte-like cells, forming a polarized monolayer. | The primary in vitro model for predicting human intestinal absorption and permeability [19]. |
| Transwell Plates | Multi-well plates with permeable membrane inserts that allow for compartmentalized cell culture. | Used in Caco-2 assays to separate apical and basolateral compartments for permeability measurement [19]. |
| RDKit | An open-source cheminformatics toolkit. | Used to compute molecular descriptors (e.g., RDKit 2D descriptors) and generate molecular fingerprints (e.g., Morgan fingerprints) for machine learning models [19]. |
| Molecular Operating Environment (MOE) | Commercial software suite for molecular modeling and drug discovery. | Used for molecular docking studies to predict binding interactions with targets like metabolic enzymes or viral proteins [21]. |
| PharmaBench | A comprehensive, open-source benchmark set for ADMET properties with over 52,000 entries. | Serves as a high-quality, diverse dataset for training and validating in silico ADMET prediction models [22]. |
| SwissADME / pkCSM | Free web servers for predicting pharmacokinetic and toxicity properties. | Provide accessible in silico predictions for parameters like log P, solubility, and CYP inhibition during early-stage screening [18]. |
Integrated Workflows: Bridging In Silico and In Vitro Data
The most effective strategy to mitigate ADMET-related failure is an integrated workflow that iteratively cycles between computational prediction and experimental validation. This approach ensures that only the most promising compounds advance, saving time and resources. The following diagram illustrates this iterative validation cycle.
Future Perspectives in ADMET Prediction
The future of ADMET prediction lies in enhancing the accuracy and integration of models. The use of Generative AI (GenAI) for de novo molecular design is emerging, with a focus on creating "beautiful molecules" that are synthetically feasible and have optimal ADMET profiles from the outset [23]. A key challenge remains the accurate prediction of complex properties like binding affinity and toxicity when exploring novel chemical spaces [23]. Furthermore, the creation of larger, more standardized, and clinically relevant benchmark datasets, such as PharmaBench, is crucial for developing robust models [22]. The ultimate goal is a closed-loop discovery system where AI-generated molecules are rapidly synthesized and tested, with the resulting data continuously refining the predictive models, thereby accelerating the journey to safe and effective therapeutics [23] [17].
Drug-drug interactions (DDIs) represent a significant clinical challenge, potentially leading to serious adverse events, reduced treatment efficacy, and even market withdrawal of pharmaceuticals [24]. For decades, drug development programs faced practical challenges in designing and interpreting DDI studies due to differing regional guidance from major regulatory agencies including the US Food and Drug Administration (FDA), the European Medicines Agency (EMA), and Japan's Pharmaceuticals and Medical Devices Agency (PMDA) [25] [26]. The International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use (ICH) initiated the M12 guideline to address these disparities and create a single set of globally harmonized recommendations for designing, conducting, and interpreting metabolic enzyme- or transporter-mediated drug-drug interaction studies [27] [28] [26]. This harmonization aims to streamline global drug development, facilitate patient access to new therapies, and ensure consistent safety standards across regions [25].
The ICH M12 guideline, which reached its final version in 2024 after a draft release in 2022, provides a consolidated framework that supersedes previous regional guidances [27] [28]. This article examines the role of ICH M12 in achieving regulatory harmonization, with particular focus on its implications for validating in silico ADMET predictions against in vitro data—a critical component of modern drug development workflows.
Comparative Analysis of DDI Guidelines: Pre- and Post-ICH M12
Key Regional Differences Before ICH M12
Prior to the adoption of ICH M12, regional regulatory agencies maintained distinct guidelines with variations in experimental protocols, interpretation criteria, and submission requirements. These differences created complexities for sponsors seeking global approval for new therapeutic products [25] [26]. The table below summarizes the major regional guidances that ICH M12 replaces or consolidates.
Table 1: Major Regional DDI Guidances Consolidated by ICH M12
| Regulatory Agency | Previous Guidance Document | Key Characteristics | Status with ICH M12 |
|---|---|---|---|
| US FDA | In Vitro and Clinical DDI Guidance (2020) | Separate documents for in vitro and clinical studies; specific FDA recommendations | Replaced by ICH M12 [25] |
| European Medicines Agency (EMA) | Guideline on Investigation of Drug Interactions - Revision 1 (2013) | Comprehensive coverage including GI-mediated interactions | Superseded by ICH M12 [28] |
| Japan PMDA | DDI Guidance (2018) | Specific requirements for Japanese submissions | Replaced by harmonized approach [25] |
Substantive Changes in ICH M12
ICH M12 introduces significant changes to DDI evaluation criteria that affect both experimental design and interpretation. These modifications create a unified standard for assessing interaction potential across regulatory jurisdictions. The following table compares key evaluation parameters between previous approaches and the new ICH M12 standards.
Table 2: Comparison of Key DDI Evaluation Parameters Before and After ICH M12
| Evaluation Area | Previous Regional Variations | ICH M12 Harmonized Approach | Impact on DDI Assessment |
|---|---|---|---|
| Protein Binding | Differing recommendations between FDA and EMA on using unbound fraction [29] | Use of unbound human plasma fraction <0.01 allowed with proper methodology [30] [29] | May decrease predicted interaction risk for highly bound compounds [29] |
| CYP Induction Concentration | FDA used 50× Cmax,u; EMA used lower multiples [30] | Standardized to 50× Cmax,u for induction risk assessment [30] [29] | More consistent induction potential evaluation |
| Time-Dependent Inhibition (TDI) | Primarily dilution assays recommended [29] | Both dilution and non-dilution methods accepted [30] [29] | Increased methodological flexibility |
| Metabolite as Inhibitor | Threshold differences between regions [25] | Consistent threshold: AUCmetab ≥25% of AUCparent and ≥10% of drug-related material [25] [30] | Standardized metabolite DDI assessment |
| Transporter Inhibition Cut-offs | Different R-values between FDA and EMA [30] | Harmonized cut-off values for positive signals [30] | Consistent transporter DDI interpretation |
| UGT Enzyme Evaluation | Minimal guidance in FDA's previous guidance [25] | Detailed recommendations with list of substrates and inhibitors [25] | Enhanced evaluation of glucuronidation interactions |
Experimental Protocols and Methodological Standards
In Vitro DDI Evaluation Framework
ICH M12 provides detailed methodological recommendations for in vitro DDI studies that support the validation of in silico ADMET predictions. These protocols establish standardized conditions for assessing enzyme- and transporter-mediated interactions.
Enzyme-Mediated DDI Assessments:
- Reaction Phenotyping: Experiments should identify enzymes contributing ≥25% to drug elimination [24]. Cytochrome P450 enzymes (CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A) must be routinely evaluated using in vitro reaction phenotyping, preferably before Phase 1 trials [24] [30].
- CYP Inhibition Studies: Both direct and time-dependent inhibition should be tested for all major CYP enzymes using pooled human liver microsomes (HLM), pooled hepatocytes, or microsomes from recombinant systems [24]. Determination of Ki or IC50 should use several concentrations of the investigational drug relevant to clinical exposure [24].
- CYP Induction Studies: Testing for CYP1A2, CYP2B6, and CYP3A4 induction is recommended using human hepatocytes from a minimum of three individual donors [24]. The preferred readout (except for CYP2C19) is changes in CYP450 mRNA levels [24].
Transporter-Mediated DDI Assessments:
- Efflux Transporters: In vitro P-gp and BCRP substrate and inhibition data are typically expected in regulatory submissions [24]. Bidirectional transport assays with cell-based systems are recommended, especially for drugs where biliary excretion is a major elimination pathway or when the pharmacological target is in the brain [24].
- Hepatic Uptake Transporters: Evaluation as a substrate for OATP1B1 and OATP1B3 is recommended if hepatic metabolism or biliary excretion accounts for ≥25% of drug elimination or if the pharmacological target is in the liver [24] [30].
- Renal Transporters: Assessment of OAT1, OAT3, OCT2, MATE1, and MATE2-K is recommended if a drug demonstrates renal toxicity or if renal active secretion accounts for ≥25% of systemic clearance [24].
Timing of DDI Evaluations in Drug Development
ICH M12 provides clearer recommendations on when to conduct specific DDI assessments throughout the drug development continuum [25] [24]:
Table 3: Recommended Timing for DDI Assessments in Drug Development
| Development Stage | Required DDI Assessments | Purpose and Rationale |
|---|---|---|
| Pre-Phase 1 | In vitro reaction phenotyping (enzymes) [24] [30] | Identify major metabolic pathways to inform initial clinical trial design and safety monitoring |
| Pre-Phase 1 | In vitro precipitant effects on CYP enzymes and transporters [30] | Understand potential perpetrator effects to guide exclusion criteria for concomitant medications |
| During Clinical Development | In vitro interactions for major/active metabolites [30] | Characterize metabolite DDI potential once human metabolic profile is established |
| Before Phase 3 | Human absorption, metabolism, and excretion (hAME) study results [30] | Comprehensive understanding of elimination pathways to inform final DDI strategy |
| Before Phase 3 | Clinical DDI studies based on integrated in vitro and clinical data [24] | Final confirmation of DDI risk to inform product labeling |
Model-Informed Drug Development Approaches
ICH M12 explicitly recognizes the value of model-based approaches for DDI evaluation [28] [31] [30]. The guideline describes the application of both mechanistic static models (MSM) and physiologically based pharmacokinetic (PBPK) modeling in various contexts:
- Mechanistic Static Models: These can be used to extrapolate victim DDI results with CYP or transporter inhibitors to "less potent" inhibitors, allowing for a semi-quantitative approach based on justifications and sensitivity analyses [30].
- PBPK Modeling: Recommended for informing DDI strategy and study design, translating pharmacogenomic effects to object DDIs, evaluating complex DDIs, DDIs in specific populations, replacing studies of staggered dosing, supporting evaluations of drugs with long half-lives, and leveraging endogenous biomarker data [31] [30].
- Endogenous Biomarkers: ICH M12 endorses the use of biomarkers such as plasma coproporphyrin I (for hepatic OATP1B1/3), plasma and urine N-methylnicotinamide and N-methyladenosine (for renal OCT2, MATE1, MATE2K), and plasma 4β-hydroxycholesterol/cholesterol ratio (for CYP3A) [30]. These biomarkers can be leveraged in PBPK models to strengthen DDI predictions [30].
The following diagram illustrates the integrated experimental and computational workflow for DDI assessment under ICH M12:
Diagram 1: Integrated DDI Assessment Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
Implementation of ICH M12-compliant DDI assessments requires specific research reagents and experimental systems. The following table details essential materials for conducting these evaluations.
Table 4: Essential Research Reagents for ICH M12-Compliant DDI Assessments
| Reagent/System | Function in DDI Assessment | Specific Application Examples |
|---|---|---|
| Pooled Human Liver Microsomes (HLM) | Evaluation of metabolic stability and enzyme inhibition potential [24] | CYP reaction phenotyping; reversible inhibition assays |
| Transfected Cell Lines | Transporter substrate and inhibition assays [24] | P-gp, BCRP, OATP1B1, OATP1B3, OATs, OCTs, MATEs evaluation |
| Cryopreserved Human Hepatocytes | Assessment of enzyme induction potential and metabolic clearance [24] | CYP1A2, CYP2B6, CYP3A4 induction assays; metabolite identification |
| Recombinant CYP Enzymes | Reaction phenotyping to identify specific enzymes involved in metabolism [24] | Determination of enzyme-specific contribution to total metabolism |
| Specific Probe Substrates | Evaluation of enzyme and transporter inhibition potential [24] [30] | Quantitative assessment of inhibitory potency (IC50, Ki) |
| Validated Chemical Inhibitors | Selective inhibition of specific enzymes or transporters in phenotyping studies [24] | Identification of contribution of specific pathways to total clearance |
Implications for In Silico ADMET Prediction Validation
The harmonization achieved through ICH M12 has significant implications for validating in silico ADMET predictions, creating more standardized datasets for model training and verification.
Standardized Data for Model Development
The consistent experimental protocols and interpretation criteria established by ICH M12 generate standardized datasets that enhance the reliability of in silico ADMET models in several key areas:
- Protein Binding Considerations: ICH M12's guidance on using experimentally measured fraction unbound for drugs with >99% protein binding enables more accurate prediction of unbound drug concentrations, a critical parameter for DDI risk assessment [31] [30] [29].
- Transporter DDI Prediction: Harmonized cut-off values for transporter inhibition create consistent thresholds for validating in silico predictions of transporter-mediated DDIs [30].
- Metabolite DDI Assessment: Standardized criteria for when to evaluate metabolites as substrates, inhibitors, or inducers provide clear decision trees for in silico model development [25] [30].
Integrated Computational-Experimental Workflows
ICH M12's recognition of model-informed drug development approaches supports the integration of in silico predictions with experimental data throughout the drug development process. The following diagram illustrates this integrated workflow for enzyme-mediated DDI assessment:
Diagram 2: Computational-Experimental DDI Workflow
The ICH M12 guideline represents a significant achievement in global regulatory harmonization, establishing consistent standards for DDI assessment that transcend previous regional differences. By providing unified recommendations for experimental design, methodology, and data interpretation, ICH M12 enables more efficient global drug development while maintaining rigorous safety standards.
For researchers focused on validating in silico ADMET predictions, ICH M12 creates a foundation of standardized experimental data that enhances model training and verification. The explicit recognition of model-informed drug development approaches within the guideline facilitates the integration of computational predictions with experimental verification throughout the drug development process.
As the pharmaceutical industry transitions to ICH M12 standards, the harmonized framework will likely accelerate the adoption and refinement of in silico ADMET prediction methods, ultimately contributing to more efficient drug development and improved patient safety through better prediction and management of drug interactions.
In silico models, particularly for predicting the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) of drug candidates, have become indispensable tools in modern drug discovery, offering a scalable and efficient alternative to resource-intensive traditional methods [32] [33]. These computer-based simulations allow for the high-throughput screening of compounds, significantly accelerating the lead optimization phase [33] [34]. The ultimate goal is to mitigate the high attrition rates in clinical development, where poor pharmacokinetics and unforeseen toxicity remain major causes of failure [33]. However, the reliability of these in silico predictions hinges on their validation against experimental data, with in vitro assays serving as a crucial benchmark for establishing real-world biological relevance before proceeding to complex and costly in vivo studies [35] [36]. This guide objectively compares the performance of various in silico approaches against in vitro data, examining the key challenges of data quality and model generalization that define the current landscape.
Performance Comparison: In Silico Predictions vs. In Vitro Data
The following tables summarize quantitative performance data and core challenges when comparing in silico model predictions with experimental in vitro results.
Table 1: Quantitative Performance Benchmarks of In Silico Models Against In Vitro Data
| ADMET Property | Typical In Vitro Assay | High-Performing In Silico Models | Reported Performance (AUC/R²/Accuracy) | Key Limitations vs. In Vitro |
|---|---|---|---|---|
| Absorption (Permeability) | Caco-2 cell assay [33] | Graph Neural Networks (GNNs), Ensemble Methods [33] | R²: ~0.67-0.68 (on analogous endpoints) [37] | Struggles with active transport mechanisms (e.g., P-gp) not fully captured by structure [33] |
| Metabolism (CYP Inhibition) | Human liver microsomes, recombinant enzymes [33] | Multitask Deep Learning, XGBoost [33] | High AUC values reported for major CYP isoforms [33] | Predicts potential, not actual metabolic rate; misses novel metabolites [33] |
| Toxicity (hERG) | hERG potassium channel assay [33] | Machine Learning on molecular descriptors [32] [33] | Accuracy often >70% in research settings [32] | High false-negative risk for structurally novel scaffolds; lacks organ-level context [33] |
| Blastocyst Formation (IVF) | Embryo morphology assessment [38] | LightGBM, XGBoost, SVM [37] | R²: 0.673–0.676; Accuracy: 0.675–0.71 [37] | Model may underestimate yields in poor-prognosis subgroups [37] |
Table 2: Core Data Quality and Generalization Challenges
| Challenge Category | Impact on In Silico Model Performance | Manifestation in In Vitro Validation |
|---|---|---|
| Data Quality & Availability | Model accuracy is highly dependent on the quality, size, and chemical diversity of the training data [32] [34]. | Predictions are unreliable for chemical spaces not represented in the training set, leading to high error rates when tested with novel compounds in vitro [33]. |
| Algorithmic Limitations & Black-Box Nature | Deep learning models, while powerful, often lack interpretability, making it difficult to understand the rationale behind a prediction [33]. | Difficult for scientists to trust or troubleshoot mismatches between in silico and in vitro results without mechanistic insights [33]. |
| Experimental Variability & Biological Complexity | Inconsistencies in experimental protocols and biological noise in the in vitro data used for training confound model learning [38]. | Models trained on one lab's in vitro data may not generalize to another lab's data due to differences in assay conditions or cell lines [38]. |
| Contextual Oversimplification | Models predict based on molecular structure alone, missing the integrated physiology of a living system [35] [36]. | A compound predicted to have high permeability in silico may show poor absorption in vitro due to efflux transporters or metabolism not modeled [33]. |
Experimental Protocols for Model Validation
A rigorous and transparent methodology is essential for the meaningful validation of in silico ADMET predictions against in vitro benchmarks. The following workflow outlines a standardized protocol for this process.
Detailed Methodological Steps
Compound Selection and Curation: Select a diverse and chemically representative set of drug candidates not used in the model's training. Curate structures using standardized formats (e.g., SMILES) and ensure purity is verified for in vitro testing [34].
In Silico Prediction Execution: Apply the trained machine learning model (e.g., GNN, LightGBM) to generate predictions for the specific ADMET endpoint (e.g., Caco-2 permeability, hERG inhibition). All predictions and associated confidence scores should be documented before in vitro testing [33] [34].
Parallel In Vitro Assay Performance: Conduct the corresponding gold-standard in vitro assay (e.g., Caco-2 for permeability, hERG patch clamp for toxicity) following strict, standardized operating procedures (SOPs) to minimize experimental variability. Assays should be performed in replicates, and raw data should be recorded with metadata on assay conditions [33] [38].
Data Integration and Statistical Comparison: Integrate the in silico predictions and in vitro results into a unified dataset. Calculate a suite of performance metrics to evaluate the agreement, including:
- Discrimination: Area Under the Receiver Operating Characteristic Curve (AUC-ROC) for classification tasks.
- Accuracy & Error: R-squared (R²) and Mean Absolute Error (MAE) for regression tasks [37].
- Calibration: Analysis of how well the predicted probabilities match the observed frequencies in vitro [38].
Discrepancy Analysis and Model Iteration: Systematically investigate compounds where major discrepancies occur between prediction and assay results. This analysis can reveal model blind spots and inform the refinement of the training set or algorithm, leading to model retraining for improved generalizability [33] [34].
The Scientist's Toolkit: Essential Research Reagent Solutions
Successful validation relies on specific, high-quality reagents and tools. The following table details essential materials for featured ADMET validation workflows.
Table 3: Key Research Reagents and Tools for ADMET Validation
| Reagent/Tool Name | Function in Workflow | Specific Application Example |
|---|---|---|
| Caco-2 Cell Line | A human colon adenocarcinoma cell line used as an in vitro model of the human intestinal mucosa to predict drug absorption [33]. | Measuring apparent permeability (Papp) of drug candidates for comparison with in silico absorption predictions [33]. |
| hERG-Expressing Cell Line | Cell lines (e.g., HEK293) stably expressing the human Ether-à-go-go-Related Gene potassium channel. | In vitro patch-clamp or flux assays to assess compound risk for Torsades de Pointes cardiac arrhythmia, validating in silico toxicity alerts [33]. |
| Human Liver Microsomes (HLM) | Subcellular fractions containing cytochrome P450 (CYP) enzymes and other drug-metabolizing enzymes. | Incubated with a drug candidate to identify major metabolites and calculate metabolic stability (e.g., half-life), grounding truth for in silico metabolism models [33]. |
| Standardized Molecular Descriptors | Numerical representations of chemical structures (e.g., ECFP, molecular weight, logP) used as input for ML models. | Enable quantitative structure-activity relationship (QSAR) modeling for ADMET endpoints. Critical for model interoperability and performance [32] [34]. |
| Graph Neural Network (GNN) Framework | A class of deep learning algorithms that operate directly on molecular graph structures. | Captures complex structure-property relationships for ADMET endpoints, often leading to higher predictive accuracy compared to traditional descriptors [33]. |
The current landscape of in silico ADMET prediction is defined by a tension between immense promise and persistent challenges. While advanced machine learning models like graph neural networks and ensemble methods increasingly demonstrate robust performance, their utility in de-risking drug development is ultimately constrained by the quality of the underlying data and their ability to generalize beyond their training sets. The critical practice of rigorous, multi-faceted validation against standardized in vitro assays remains the cornerstone for building trust in these in silico tools. Future progress hinges on the generation of higher-quality, more comprehensive experimental data, the development of more interpretable and biologically integrated models, and a continued commitment to transparent and standardized benchmarking. By systematically addressing these challenges of data quality and model generalization, the field can fully realize the potential of in silico methods to accelerate the delivery of safer and more effective therapeutics.
Building and Applying Integrated In Silico and In Vitro Workflows
In modern drug development, the assessment of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties represents a critical gatekeeper determining candidate success or failure. Historically, poor ADMET profiles have been responsible for approximately 40-60% of clinical trial failures, creating compelling economic and ethical imperatives for earlier, more reliable prediction [39]. The pharmaceutical industry has consequently shifted toward extensive ADMET screening earlier in the discovery process to identify and eliminate problematic compounds before they enter costly development phases [40].
In silico (computational) methods have emerged as powerful tools addressing this challenge, offering rapid, cost-effective alternatives to expensive and time-consuming experimental testing. These approaches eliminate the need for physical samples and laboratory facilities while providing critical insights into compound behavior [40]. This guide examines the evolving landscape of in silico ADMET tools, from established Quantitative Structure-Activity Relationship (QSAR) methods to advanced machine learning (ML) algorithms and sophisticated Physiologically-Based Pharmacokinetic (PBPK) modeling platforms, with particular emphasis on validation against experimental data.
Fundamental Methods: QSAR and Molecular Modeling
QSAR Foundations and Applications
Quantitative Structure-Activity Relationship (QSAR) modeling represents the foundational approach for predicting chemical properties from molecular structure. QSAR models correlate structural descriptors of compounds with their biological activities or physicochemical properties through statistical methods, enabling property prediction for novel compounds based on their structural features [39].
The predictive performance of QSAR models is highly dependent on the quality and diversity of their training data and the relevance of selected molecular descriptors. Recent benchmarking studies of twelve QSAR software tools demonstrated adequate predictive performance for many physicochemical properties (average R² = 0.717), with slightly lower performance for toxicokinetic properties (average R² = 0.639 for regression models) [39]. These tools have become increasingly sophisticated, with applications ranging from predicting basic physicochemical properties like solubility and lipophilicity to complex metabolic stability and transporter affinity.
Molecular Modeling Techniques
Beyond traditional QSAR, more computationally intensive molecular modeling methods provide atomic-level insights into ADMET processes:
Quantum Mechanics (QM) and Molecular Mechanics (MM): QM calculations explore electronic structure properties that influence chemical reactivity and metabolic transformations, particularly valuable for understanding cytochrome P450 metabolism mechanisms [40]. The hybrid QM/MM approach combines accuracy of QM for reaction centers with efficiency of MM for protein environments.
Molecular Dynamics (MD) Simulations: MD tracks atom movements over time, revealing binding/unbinding processes, conformational changes, and passive membrane permeability that directly influence absorption and distribution properties [11].
Molecular Docking: This technique predicts how small molecules bind to protein targets like metabolic enzymes or transporters, providing insights into substrate specificity and inhibition potential [41].
Table 1: Molecular Modeling Methods for ADMET Prediction
| Method | Key Applications in ADMET | Computational Cost | Key Insights Provided |
|---|---|---|---|
| QSAR | High-throughput property prediction | Low | Structure-property relationships across compound libraries |
| Molecular Docking | Metabolic enzyme binding, transporter interactions | Medium | Binding modes, affinity estimates, molecular interactions |
| MD Simulations | Membrane permeability, conformational changes | High | Time-dependent behavior, free energy calculations |
| QM/MM | Metabolic reaction pathways, reactivity | Very High | Electronic structure effects, reaction mechanisms |
Machine Learning and Deep Learning Approaches
Algorithmic Advances in ADMET Prediction
Machine learning (ML) has dramatically expanded capabilities for ADMET prediction, moving beyond traditional QSAR's linear assumptions to capture complex, nonlinear relationships in chemical data. Commonly employed algorithms include Random Forest (RF), Support Vector Machines (SVM), XGBoost, and Gradient Boosted Machines (GBM), each with strengths for different prediction tasks [42] [19].
More recently, deep learning (DL) approaches using Graph Neural Networks (GNNs) and Message-Passing Neural Networks (MPNNs) have demonstrated state-of-the-art performance by directly learning from molecular graph representations rather than pre-defined descriptors [42] [19]. For example, Directed-MPNN (D-MPNN) has shown particular promise in molecular property prediction by operating on the graph structure of molecules and passing messages through edge-dependent neural networks [42].
Case Study: Caco-2 Permeability Prediction
Caco-2 cell monolayer permeability represents a critical parameter for predicting intestinal absorption of oral drugs. Traditional experimental assessment requires 7-21 days for cell differentiation, creating bottlenecks in early discovery [19]. Machine learning models address this limitation through quantitative prediction from chemical structure alone.
A comprehensive benchmarking study evaluated multiple ML algorithms using a large dataset of 5,654 curated Caco-2 permeability measurements [19]. The research compared four machine learning methods (XGBoost, RF, GBM, SVM) and two deep learning approaches (D-MPNN and CombinedNet) using diverse molecular representations including Morgan fingerprints, RDKit 2D descriptors, and molecular graphs. The study found that XGBoost generally provided superior predictions, with model performance robust across different dataset splits [19].
Table 2: Performance Comparison of ML Algorithms for Caco-2 Permeability Prediction
| Algorithm | Molecular Representation | R² | RMSE | Key Advantages |
|---|---|---|---|---|
| XGBoost | Morgan fingerprints + RDKit 2D | 0.81 | 0.31 | Best overall performance, handling of non-linear relationships |
| Random Forest | Morgan fingerprints + RDKit 2D | 0.78 | 0.33 | Robust to outliers, feature importance analysis |
| GBM | Morgan fingerprints + RDKit 2D | 0.79 | 0.32 | Good balance of performance and training speed |
| D-MPNN | Molecular graphs | 0.76 | 0.35 | Automatic feature learning, no descriptor engineering required |
| SVM | Morgan fingerprints + RDKit 2D | 0.72 | 0.38 | Effective in high-dimensional spaces |
The transferability of models trained on public data to industrial settings was also investigated using an internal pharmaceutical company dataset. Results demonstrated that boosting models retained reasonable predictive performance when applied to industry compounds, though some performance degradation highlighted the importance of domain applicability [19].
PBPK Modeling: Integrating Physiology and Mechanism
Principles and Applications of PBPK Modeling
Physiologically-Based Pharmacokinetic (PBPK) modeling represents a mechanistic approach that simulates drug disposition by incorporating physiological parameters (organ volumes, blood flows), drug-specific properties (lipophilicity, permeability, binding), and system-specific characteristics (enzyme/transporter abundances) [43] [44]. Unlike purely empirical models, PBPK models maintain direct physiological relevance, enabling prediction of drug concentrations in specific tissues and extrapolation to special populations [43].
PBPK modeling has proven particularly valuable in scenarios where clinical data are limited or difficult to obtain due to ethical constraints, such as in pediatric or geriatric populations, pregnant women, and patients with organ impairments [44]. These models can also predict variations in drug metabolism resulting from genetic polymorphisms (e.g., in CYP2D6, CYP2C9, CYP2C19), age-related physiological changes, and disease states [44].
Current Challenges and Limitations
Despite their power, traditional PBPK models face several significant challenges:
Parameter Uncertainty: PBPK models depend on numerous physiological and drug-specific parameters, many of which have substantial uncertainty or inter-individual variability [43]. For example, values for lymph flow rates used in antibody PBPK models vary by two orders of magnitude across different publications [43].
Model Complexity: Comprehensive PBPK models can become extraordinarily complex. A full PBPK model for a therapeutic antibody may require knowledge of over a dozen parameters per tissue compartment, with extrapolation to multiple organs dramatically increasing the parameter estimation challenge [43].
Limited Data Availability: Local drug concentrations in different cells and tissues are rarely available for model verification, creating validation challenges [43].
Extension to Novel Formulations: Adapting PBPK models for new drug delivery systems (e.g., nanoparticles) requires accounting for entirely new processes like uptake by the mononuclear phagocytic system, with additional parameters that are often poorly characterized [43].
Hybrid Approaches: Integrating Machine Learning with PBPK Modeling
The ML-PBPK Framework
Recent advances have focused on integrating machine learning with PBPK modeling to overcome traditional limitations. This hybrid approach uses ML to predict critical drug-specific parameters directly from chemical structure, which are then incorporated into mechanistic PBPK frameworks [45] [42].
A landmark study developed an ML-PBPK platform that predicts human pharmacokinetics from compound structures without requiring experimental data [42]. The approach used machine learning models to predict three key parameters: plasma protein fraction unbound (fup), Caco-2 cell permeability, and total plasma clearance (CLt). These ML-predicted parameters were then used as inputs for a whole-body PBPK model encompassing 14 tissues [42].
The results demonstrated that the ML-PBPK model predicted the area under the concentration-time curve (AUC) with 65.0% accuracy within a 2-fold range, significantly outperforming PBPK models using traditional in vitro inputs (47.5% accuracy within 2-fold) [42]. This represents a substantial improvement in predictive performance while simultaneously reducing experimental requirements.
Diagram 1: ML-PBPK Integrated Modeling Workflow
Case Study: AI-PBPK for Aldosterone Synthase Inhibitors
A specialized AI-PBPK model was developed to predict pharmacokinetic and pharmacodynamic properties of aldosterone synthase inhibitors (ASIs) during early discovery stages [45]. The model integrated machine learning with classical PBPK modeling to enable PK simulation of ASIs directly from their structural formulas.
The workflow involved:
- Inputting the compound's structural formula into the AI model to generate key ADME parameters and physicochemical properties
- Using these parameters in the PBPK model to predict pharmacokinetic profiles
- Developing a PD model to predict inhibition rates of aldosterone synthase and 11β-hydroxylase based on plasma free drug concentrations [45]
This approach successfully predicted PK/PD properties for multiple ASI compounds from their structural formulas alone, providing valuable reference for early lead compound screening and optimization [45]. The model demonstrated that AI-PBPK integration could significantly accelerate candidate selection while reducing resource-intensive experimental screening.
Benchmarking and Validation Frameworks
Software Tool Performance Assessment
Comprehensive benchmarking of computational tools is essential for assessing their real-world predictive performance. A recent evaluation of twelve QSAR software tools across 41 validation datasets for 17 physicochemical and toxicokinetic properties provided valuable insights into the current state of computational ADMET prediction [39].
Key findings included:
- Models for physicochemical properties generally outperformed those for toxicokinetic properties
- Several tools exhibited good predictivity across different properties and were identified as recurring optimal choices
- The importance of applicability domain assessment for identifying reliable predictions
- Significant performance variation across different chemical classes and property types
Table 3: Performance Summary of Selected ADMET Prediction Tools
| Software Tool | Key Features | Supported Properties | Performance Notes |
|---|---|---|---|
| OPERA | Open-source QSAR models, applicability domain assessment | PC properties, environmental fate, toxicity | Good predictivity for logP, water solubility |
| SwissADME | Web-based, user-friendly interface | Physicochemical properties, drug-likeness, pharmacokinetics | Free tool with comprehensive ADME profiling |
| ADMETlab 3.0 | Platform with multiple prediction modules | Comprehensive ADMET endpoints | High efficiency for large-scale screening |
| B2O Simulator | AI-PBPK integrated platform | PK/PD prediction from structure | Specialized for pharmacokinetic simulation |
Emerging Benchmark Datasets
The development of robust benchmark datasets like PharmaBench addresses critical limitations in previous ADMET datasets, which were often too small or unrepresentative of drug discovery compounds [22]. PharmaBench comprises eleven ADMET datasets with 52,482 entries, significantly larger and more diverse than previous resources.
This benchmark was constructed using a novel multi-agent data mining system based on Large Language Models (LLMs) that effectively identifies experimental conditions within 14,401 bioassays, enabling proper merging of entries from different sources [22]. Such comprehensive, well-curated benchmarks are essential for rigorous tool validation and development of next-generation predictive models.
Table 4: Key Research Reagents and Computational Resources for ADMET Prediction
| Resource | Type | Primary Function | Application Context |
|---|---|---|---|
| Caco-2 Cell Line | In vitro system | Intestinal permeability assessment | Gold standard for absorption prediction; training data for ML models |
| Human Hepatocytes | In vitro system | Hepatic metabolism studies | Clearance prediction, metabolite identification |
| CYP Enzymes | Recombinant enzymes | Metabolic stability screening | Reaction phenotyping, enzyme kinetics |
| RDKit | Open-source cheminformatics | Molecular descriptor calculation | Feature generation for QSAR/ML models |
| Chemprop | Deep learning package | Molecular property prediction | D-MPNN implementation for ADMET endpoints |
| PharmaBench | Curated dataset | Model training and benchmarking | Standardized evaluation across methods |
| GastroPlus | Commercial software | PBPK modeling | Mechanistic PK simulation and prediction |
| Simcyp Simulator | Commercial platform | PBPK modeling | Population-based PK prediction |
The evolution of in silico ADMET tools has progressed from isolated QSAR models to integrated workflows combining machine learning with mechanistic modeling. The most promising approaches leverage ML for parameter prediction from structure while maintaining physiological relevance through PBPK frameworks [45] [42]. This integration addresses key limitations of both methods: the black-box nature of pure ML predictions and the parameter uncertainty of traditional PBPK models.
Future directions point toward continued refinement of these hybrid models, expansion of high-quality benchmarking datasets, and improved applicability domain characterization for reliable prediction. As these tools mature, they offer the potential to significantly reduce animal testing, accelerate candidate selection, and improve clinical success rates through earlier, more accurate ADMET profiling [43] [42].
Diagram 2: Integrated ADMET Prediction and Validation Workflow
Designing Physiologically Relevant In Vitro Assays for Key ADMET Endpoints
Accurately predicting the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties of drug candidates remains a fundamental challenge in modern drug discovery. The pharmaceutical industry faces significant setbacks when promising compounds fail during development due to suboptimal ADMET characteristics, contributing to substantial financial losses and extended development timelines [1] [46]. While in silico approaches have emerged as valuable tools for early screening, their predictive accuracy ultimately depends on validation against biologically relevant experimental data [47]. This creates an pressing need for physiologically relevant in vitro assays that can bridge the gap between computational predictions and in vivo outcomes.
The transition from traditional nominal concentration reporting to free concentration measurements represents a paradigm shift in assay design. Nominal concentration, defined as the total mass of a chemical added to a defined volume of exposure medium, has been criticized for not accurately reflecting in vivo biologically effective doses due to differences in biokinetics [48]. Consequently, the freely dissolved concentration in media is increasingly recognized as a more appropriate metric for comparisons with freely dissolved concentrations in plasma from in vivo testing [48]. This review systematically compares current approaches for designing physiologically relevant in vitro assays, with particular emphasis on mass balance models, experimental protocols, and their validation within the broader context of computational ADMET prediction.
Comparative Analysis of In Vitro Mass Balance Models
Model Architectures and Applicability Domains
In vitro mass balance models have been developed to characterize chemical distribution and predict cellular and free chemical concentrations, addressing the limitations of nominal concentration measurements [48]. These models simulate chemical partitioning across various compartments, including media constituents, extracellular matrices, test system materials, intracellular accumulation, volatilization, and abiotic degradation [48]. A recent comprehensive evaluation compared four established models with broad applicability to chemical types and in vitro systems, revealing significant differences in their approaches and performance characteristics (Table 1).
Table 1: Comparative Performance of In Vitro Mass Balance Models for Predicting Free Concentrations
| Model Reference | Applicable Chemicals | Model Type | Key Partitions Considered | Additional Factors | Prediction Accuracy |
|---|---|---|---|---|---|
| Fischer et al. | Neutral/ionized; Non-volatile | Equilibrium partitioning | Media, Cells (protein & lipid) | - | Moderate for media concentrations |
| Armitage et al. | Neutral/ionized; Volatile/Non-volatile | Equilibrium partitioning | Media, Cells, Labware, Headspace | Solubility limitations | Highest overall accuracy |
| Fisher et al. | Neutral/ionized; Volatile/Non-volatile | Time-dependent | Media, Cells (multi-compartment), Labware, Headspace | Metabolic transformation | Good for dynamic systems |
| Zaldivar-Comenges et al. | Neutral; Volatile/Non-volatile | Time-dependent | Media, Cells (multi-compartment), Labware, Headspace | Evaporation, abiotic degradation, cell growth | Limited to neutral compounds |
The Armitage model demonstrated slightly superior performance overall, particularly for predicting media free concentrations, which were generally more accurate than cellular concentration predictions across all models [48]. Through sensitivity analyses, researchers determined that chemical property-related parameters were most influential for media predictions, while cell-related parameters gained importance for cellular predictions [48]. This highlights the critical importance of accurate input parameters, especially for applications in Quantitative in vitro to in vivo extrapolation (QIVIVE).
Impact on QIVIVE Concordance
The practical utility of these mass balance models was assessed through their impact on QIVIVE accuracy for a dataset of 15 chemicals with both in vitro and regulatory in vivo points-of-departure. Interestingly, incorporating in vitro and in vivo bioavailability resulted in only modest improvements to in vitro-in vivo concordance [48]. This suggests that while mass balance models improve accuracy for predicting free concentrations, other factors likely contribute to the remaining discordance between in vitro and in vivo systems.
Based on these comprehensive evaluations, researchers concluded that a reasonable first-line approach for incorporating in vitro bioavailability into QIVIVE would be to use the Armitage model to predict media concentrations, while prioritizing accurate chemical property data as input parameters [48]. This recommendation balances predictive performance with practical implementation considerations.
Experimental Protocols for Key ADMET Endpoints
Free Concentration Measurement Methodology
Accurately determining free concentrations in vitro requires careful experimental design and execution. The following protocol outlines the key steps for measuring free media concentrations using mass balance principles:
System Characterization: Precisely quantify the volumes and compositions of all assay compartments, including media volume, cell volume and lipid/protein content, labware polymer type and surface area, and headspace volume [48].
Chemical Property Determination: Measure or calculate key chemical parameters including logP, pKa, molecular weight, melting point, and solubility limits in the specific test medium [48].
Distribution Experiment: Expose the in vitro system to the test compound at relevant concentrations, maintaining controlled environmental conditions (temperature, humidity, CO₂).
Sampling and Analysis: At predetermined timepoints, collect media samples and process them using appropriate methods (e.g., ultrafiltration, solid-phase microextraction) to measure free concentrations [48].
Mass Balance Verification: Confirm mass balance recovery by quantifying compound in all compartments (media, cells, labware, headspace) to identify potential loss pathways.
Model Application: Input experimental parameters into selected mass balance model to predict free concentrations and compare with measured values for validation.
This methodology enables researchers to move beyond nominal concentrations and obtain physiologically more relevant exposure metrics for QIVIVE modeling.
Protocol Standardization Challenges
A significant challenge in ADMET assay design involves the standardization of experimental protocols across different laboratories and platforms. Recent research has highlighted substantial variability in experimental conditions for even fundamental properties like aqueous solubility, which can be influenced by factors such as buffer composition, pH levels, and experimental procedures [22]. This variability complicates the integration of data from different sources and underscores the need for careful documentation of experimental parameters when designing assays for validation of in silico predictions.
Benchmarking and Validation Frameworks
PharmaBench: A Comprehensive ADMET Benchmark
The development of robust benchmarking datasets has emerged as a critical component for validating both in silico predictions and experimental assays. PharmaBench represents a significant advancement in this area, comprising eleven ADMET datasets with 52,482 entries designed specifically for evaluating predictive models in drug discovery contexts [22]. This benchmark addresses key limitations of previous datasets, including insufficient size and poor representation of compounds relevant to drug discovery projects.
Table 2: Key ADMET Endpoints Covered in PharmaBench and Experimental Considerations
| ADMET Category | Specific Endpoints | Critical Experimental Conditions | Physiological Relevance Considerations |
|---|---|---|---|
| Absorption | Caco-2 permeability, Pgp inhibition | Buffer pH, cell passage number, incubation time | Gastrointestinal pH gradients, transporter expression |
| Distribution | Plasma protein binding, blood-brain barrier penetration | Plasma source, species, temperature | Species differences in protein binding, endothelial cell models |
| Metabolism | CYP450 inhibition, metabolic stability | Enzyme source, cofactor concentrations, incubation time | Enzyme polymorphisms, non-CYP metabolism pathways |
| Excretion | Renal clearance, biliary excretion | Model system (in vitro, in vivo), species | Transporters involved in elimination, species differences |
| Toxicity | hERG inhibition, hepatotoxicity, mutagenicity | Assay format, exposure duration, endpoint measurement | Chronic vs acute exposure, metabolic activation |
PharmaBench was constructed using an innovative multi-agent Large Language Model system that extracted experimental conditions from 14,401 bioassays, enabling more precise standardization and filtering of data [22]. This approach facilitates the merging of entries from different sources while accounting for critical experimental variables that influence results.
Validation Against Regulatory Standards
Assay design must also consider regulatory requirements for ADMET evaluation. Regulatory agencies including the FDA and EMA require comprehensive ADMET assessment of drug candidates to reduce late-stage failure risks [46]. Specific expectations include:
- CYP450 inhibition and induction studies for metabolic interaction assessment
- hERG assays for cardiotoxicity risk identification
- Liver safety evaluation as part of early screening [46]
The FDA has recently outlined plans to phase out animal testing requirements in certain cases, formally including AI-based toxicity models and human organoid assays under its New Approach Methodologies framework [46]. This regulatory evolution creates opportunities for more physiologically relevant in vitro systems but necessitates rigorous validation against established standards.
Visualization of Experimental Workflows and Model Relationships
Mass Balance Model Selection Workflow
Mass Balance Model Selection
Integrated In Vitro - In Silico ADMET Validation Framework
In Vitro-In Silico Validation Framework
The Scientist's Toolkit: Essential Research Reagents and Materials
Table 3: Essential Research Reagents for Physiologically Relevant ADMET Assays
| Reagent/Material | Function | Key Considerations | Representative Examples |
|---|---|---|---|
| Mass balance model software | Predict free concentrations from nominal doses | Compatibility with chemical types and assay format | Armitage model, Fisher model, Zaldivar-Comenges model |
| Bio-relevant media | Provide physiologically relevant protein/lipid content | Species relevance, protein concentration | Serum-containing media, protein-fortified buffers |
| Metabolically competent cells | Incorporate metabolic capacity for clearance prediction | Enzyme expression levels, stability | Hepatocytes, HepaRG cells, transfected cell lines |
| Transporter-expressing systems | Assess carrier-mediated distribution | Transporter type, expression level | MDCK-MDR1, Caco-2 cells, transfected systems |
| Sorption-resistant labware | Minimize compound loss to surfaces | Polymer composition, surface treatment | Low-binding polypropylene, coated plates |
| Analytical standards | Quantify free and bound compound concentrations | Purity, stability, detection compatibility | Certified reference materials, stable isotopologs |
Designing physiologically relevant in vitro assays for ADMET endpoints requires careful consideration of mass balance principles, appropriate model selection, and standardized experimental protocols. The Armitage model currently provides the best overall performance for predicting media free concentrations, but model selection should be guided by specific assay requirements and chemical properties [48]. The emergence of comprehensive benchmarks like PharmaBench enables more rigorous validation of both experimental and computational approaches [22], while regulatory evolution creates opportunities for advanced in vitro systems to reduce reliance on animal testing [46]. As the field advances, integration of high-quality in vitro data with sophisticated in silico models will continue to improve our ability to predict human ADMET outcomes, ultimately enhancing the efficiency and success rate of drug development.
Leveraging Large-Scale Benchmark Sets like PharmaBench for Model Training
In modern drug discovery, the accurate prediction of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties has become a critical determinant of clinical success. Traditional experimental methods for ADMET assessment, while reliable, are notoriously resource-intensive and time-consuming, creating bottlenecks in the drug development pipeline [33]. Consequently, the pharmaceutical industry has increasingly turned to in silico approaches to enable early-stage screening of drug candidates with suboptimal pharmacokinetic profiles, thereby reducing late-stage attrition rates [1]. The development of robust computational models, however, is heavily dependent on access to high-quality, large-scale benchmark datasets that adequately represent the chemical space of interest in drug discovery projects.
The limitations of historical benchmark sets have been significant – many were constrained by small dataset sizes and lacked representation of compounds actually used in industrial drug discovery pipelines [22]. For instance, previous benchmarks often included compounds with molecular weights averaging only 203.9 Dalton, whereas typical drug discovery projects involve compounds ranging from 300 to 800 Dalton [22]. This representation gap fundamentally limited the utility of models trained on these datasets for real-world drug discovery applications. The emergence of next-generation benchmark sets like PharmaBench represents a transformative advancement in the field, addressing previous shortcomings through innovative data curation methodologies and substantially expanded chemical coverage [22].
Comparative Analysis of ADMET Benchmark Sets
The landscape of ADMET benchmarking resources has evolved significantly, with several major initiatives providing structured datasets for model development and evaluation. The following table summarizes the key characteristics of prominent benchmark sets used in the field.
Table 1: Comparative Analysis of Major ADMET Benchmark Sets
| Benchmark Set | Key Features | Dataset Scale | Data Sources | Primary Applications |
|---|---|---|---|---|
| PharmaBench | Multi-agent LLM system for experimental condition extraction; 11 ADMET endpoints; standardized experimental conditions | 52,482 entries from 14,401 bioassays; 156,618 raw entries processed | ChEMBL, PubChem, BindingDB, augmented with public datasets | Development of AI models for drug discovery; model evaluation across diverse ADMET properties |
| Therapeutics Data Commons (TDC) | 28 ADMET-related datasets; integrated curation from previous works | >100,000 entries | Multiple curated datasets from previous publications | Machine learning model benchmarking; therapeutic development |
| MoleculeNet | 17 datasets covering physical chemistry and physiology; broad property coverage | >700,000 compounds | Multiple public sources | Molecular machine learning benchmark; includes ADMET-related categories |
| CT-ADE | Clinical trial adverse event data; patient demographics and treatment regimens | 2,497 drugs; 168,984 drug-ADE pairs | ClinicalTrials.gov, DrugBank, MedDRA ontology | Adverse drug event prediction; clinical safety assessment |
| ADMET Benchmark Group | Framework for systematic evaluation; diverse splitting strategies | 27 properties spanning all ADME dimensions | ChEMBL, TDC, pharmaceutical company repositories | Comparative model assessment; OOD robustness evaluation |
Quantitative Comparison of Dataset Characteristics
The utility of benchmark sets for training predictive models depends heavily on specific quantitative characteristics that influence model generalizability and chemical space coverage.
Table 2: Quantitative Dataset Characteristics and Model Performance Metrics
| Benchmark Attribute | PharmaBench | Traditional Benchmarks | Impact on Model Performance |
|---|---|---|---|
| Compound Count | 52,482 curated entries | Typically hundreds to few thousand compounds | Reduces overfitting; improves generalizability |
| Molecular Weight Range | Drug-like (300-800 Dalton) | Non-drug-like (mean 203.9 Dalton) | Better translation to real drug discovery applications |
| Data Sources | 14,401 bioassays from multiple databases | Limited public sources | Increased data diversity; reduced source bias |
| Experimental Conditions | Explicitly extracted via LLM agents | Often unaccounted or not standardized | Reduces experimental noise; improves prediction accuracy |
| Assay Type Coverage | 11 key ADMET properties | Limited to few popular endpoints | Comprehensive ADMET profiling capability |
| Reported Performance Gain | Not explicitly quantified | Baseline for comparison | Up to 40-60% error reduction in related benchmarks [9] |
Experimental Protocols and Methodologies
Innovative Data Curation in PharmaBench
The creation of PharmaBench introduced a novel multi-agent Large Language Model (LLM) system that fundamentally transformed the approach to data curation for ADMET properties [22]. This system addresses the critical challenge of experimental condition variability, where identical compounds can show different results under different experimental setups (e.g., solubility varying with pH, buffer type, or experimental procedure) [22]. The data mining workflow employs three specialized agents, each with distinct functions in the data extraction and standardization process.
The Keyword Extraction Agent (KEA) identifies and summarizes key experimental conditions from assay descriptions, focusing on parameters most relevant to ADMET experiments. The Example Forming Agent (EFA) generates structured examples based on the experimental conditions identified by the KEA, creating standardized templates for data extraction. Finally, the Data Mining Agent (DMA) processes all assay descriptions to identify and extract experimental conditions using the templates created by the EFA [22]. This multi-stage approach enables the systematic transformation of unstructured experimental data from diverse sources into a standardized, machine-readable format suitable for model training.
The technical implementation of this system utilized GPT-4 as the core LLM engine, with carefully engineered prompts containing clear instructions and examples for each specific extraction task [22]. The environment for data processing was established using Python 3.12.2 within a Conda-managed virtual environment, with essential libraries including pandas, NumPy, RDKit, and scikit-learn for comprehensive cheminformatics and machine learning operations [22].
Benchmarking Protocols and Model Evaluation
Rigorous benchmarking protocols are essential for meaningful comparison of ADMET prediction models. The ADMET Benchmark Group has established comprehensive evaluation frameworks that include multiple splitting strategies to assess different aspects of model performance [49].
Scaffold-based splitting groups compounds by their molecular framework, testing a model's ability to generalize to novel chemotypes outside the training distribution. Temporal splitting arranges compounds by their date of discovery or profiling, simulating real-world scenarios where models predict properties for newly synthesized compounds. Molecular weight-constrained splitting evaluates performance across different size ranges, particularly important for assessing applicability to drug-like compounds [49].
The standard evaluation metrics encompass both regression tasks (Mean Absolute Error, Root Mean Squared Error, R²) and classification tasks (Area Under ROC Curve, Area Under Precision-Recall Curve, Matthews Correlation Coefficient) [49]. These multiple metrics provide a comprehensive view of model performance across different aspects of predictive accuracy and reliability.
Diagram 1: PharmaBench Data Curation and Model Evaluation Workflow
Performance Comparison and Experimental Data
Model Architecture Performance
Comparative studies across benchmark datasets have revealed clear performance patterns among different machine learning architectures. The ADMET Benchmark Group has systematically evaluated diverse model classes, from classical machine learning to advanced deep learning approaches [49].
Table 3: Model Architecture Performance Across ADMET Benchmarks
| Model Class | Feature Modalities | Key Strengths | Reported Performance | Limitations |
|---|---|---|---|---|
| Random Forest / GBDT | ECFP, Avalon, ErG, RDKit descriptors | State-of-the-art on several ADMET tasks; computationally efficient | Competitive performance across multiple endpoints [49] | Limited extrapolation to novel scaffolds |
| Graph Neural Networks (GAT) | Atom/bond graph representations | Best OOD generalization; robust on external data | Superior performance on novel chemical series [49] | Higher computational requirements; data hungry |
| Multimodal Approaches | Graph + molecular image fusion | Combines local and global chemical cues | Outperforms single-modal baselines [49] | Complex implementation; integration challenges |
| Foundation Models | SMILES sequences, quantum mechanical properties | Transfer learning from large unlabeled corpora | Top-1 performance in diverse benchmarks [49] | Extensive pretraining requirements |
| AutoML Frameworks | Dynamic feature selection | Automated pipeline optimization | Best performance on several datasets [49] | Limited model interpretability |
Impact of Dataset Scale on Predictive Accuracy
The relationship between dataset scale and model performance has been quantitatively demonstrated through systematic benchmarking. Recent studies indicate that models trained on larger, more diverse datasets like PharmaBench achieve significant improvements in predictive accuracy across multiple ADMET endpoints.
Federated learning initiatives that combine data from multiple pharmaceutical companies have shown that increased data diversity systematically extends model applicability domains, with performance improvements scaling with the number and diversity of participants [9]. In the Polaris ADMET Challenge, multi-task architectures trained on broader and better-curated data achieved 40-60% reductions in prediction error for critical endpoints including human and mouse liver microsomal clearance, solubility (KSOL), and permeability (MDR1-MDCKII) [9].
The performance advantage of larger benchmarks is particularly evident in out-of-distribution (OOD) scenarios, where models encounter compounds with scaffolds not represented in training data. Benchmark studies have documented that while conventional models may suffer substantial performance degradation under OOD conditions (e.g., AUC dropping from 91.97% to 83.59%), models trained on more comprehensive benchmarks demonstrate significantly better generalization [49].
Computational Tools and Databases
Successful implementation of ADMET prediction models requires access to comprehensive computational tools and data resources. The following table details essential components of the modern ADMET researcher's toolkit.
Table 4: Essential Research Resources for ADMET Prediction
| Resource Category | Specific Tools/Databases | Primary Function | Relevance to Benchmarking |
|---|---|---|---|
| Chemical Databases | ChEMBL, PubChem, BindingDB | Source of experimental bioactivity data | Foundation for benchmark curation; provides raw experimental data [22] |
| Standardized Benchmarks | PharmaBench, TDC, MoleculeNet | Curated datasets for model training/evaluation | Enable reproducible model comparison; standardized evaluation protocols [22] [49] |
| Cheminformatics Libraries | RDKit, OpenBabel, PaDEL | Molecular descriptor calculation and manipulation | Feature generation for classical ML models; molecular standardization [22] |
| Deep Learning Frameworks | PyTorch, TensorFlow, DeepChem | Implementation of neural network architectures | Enable GNNs and complex multimodal approaches [49] |
| Specialized ADMET Tools | ADMETboost, HelixADMET | Pre-trained models for specific ADMET endpoints | Baseline models for performance comparison [49] |
| Federated Learning Platforms | Apheris, MELLODDY | Cross-institutional collaboration without data sharing | Enable training on larger datasets while preserving privacy [9] |
While in silico predictions provide valuable early screening, experimental validation remains essential for confirming model predictions. Key experimental assays used for validating computational ADMET predictions include:
Caco-2 cell permeability assays for predicting intestinal absorption, human liver microsomal stability assays for metabolic clearance predictions, P-glycoprotein transporter assays for distribution and efflux potential, and plasma protein binding measurements for volume of distribution estimates [33] [50]. These experimental protocols provide the ground truth data necessary for both training computational models and validating their predictions.
For blood-brain barrier penetration, a critical distribution parameter, multiple experimental approaches are employed including in situ brain perfusion in animal models and MDCK-MDR1 cell monolayer assays [50]. The permeability values derived from these experiments, typically measured in cm/s, provide quantitative benchmarks for computational model development and validation [50].
Diagram 2: Integrated Workflow for ADMET Model Development and Validation
The advent of large-scale, carefully curated benchmark sets like PharmaBench represents a paradigm shift in ADMET prediction capabilities. By addressing critical limitations of previous datasets – including inadequate size, poor representation of drug-like compounds, and inconsistent experimental conditions – these next-generation resources enable development of more accurate and generalizable predictive models [22]. The innovative application of multi-agent LLM systems for data extraction and standardization further enhances the quality and utility of these benchmarks, providing researchers with unprecedented resources for model development [22].
Comparative analyses demonstrate that models trained on comprehensive benchmarks achieve substantial performance improvements, particularly for challenging out-of-distribution prediction tasks [49]. The integration of diverse data sources through federated learning approaches additionally expands the effective chemical space coverage, leading to models with broader applicability domains [9]. As the field continues to evolve, the systematic application of rigorous benchmarking standards and the development of increasingly sophisticated curation methodologies will be essential for further advancing predictive accuracy and clinical relevance.
The ongoing validation of in silico ADMET predictions with in vitro data remains crucial for establishing model credibility and translational potential. Through continued refinement of benchmark sets, adoption of standardized evaluation protocols, and integration of multimodal data sources, the drug discovery community moves closer to the goal of reliably predicting human pharmacokinetics and toxicity during early-stage compound design, ultimately reducing clinical attrition rates and accelerating the development of safer, more effective therapeutics.
Physiologically based pharmacokinetic (PBPK) modeling represents a mechanistic, mathematical approach that simulates the absorption, distribution, metabolism, and excretion (ADMET) of drugs in humans by integrating system-specific physiological parameters with drug-specific physicochemical and biochemical properties [44] [51]. Unlike conventional compartmental models that conceptualize the body as abstract mathematical compartments, PBPK modeling structures simulations upon a mechanism-driven paradigm, representing the body as a network of physiological compartments (e.g., liver, kidney, brain) interconnected by blood circulation [52]. This mechanistic foundation provides PBPK modeling with remarkable extrapolation capability, enabling not only the description of observed pharmacokinetic data but also the quantitative prediction of systemic and tissue-specific drug exposure under untested physiological or pathological conditions [52].
The growing adoption of PBPK modeling in drug development is evidenced by its increasing presence in regulatory submissions. According to a recent analysis of FDA-approved new drugs from 2020-2024, approximately 26.5% of submissions included PBPK models as pivotal evidence, with oncology drugs representing the largest therapeutic area (42%) [52]. This technology offers particular value in addressing ethical and practical challenges associated with clinical testing in vulnerable populations, including pregnant women, pediatric and geriatric patients, and individuals with organ impairments, where physiological and pathophysiological changes significantly alter pharmacokinetic profiles [44]. Furthermore, PBPK modeling serves as a powerful tool for predicting interindividual variability in drug responses arising from genetic polymorphisms, age-related physiological changes, and disease-induced alterations in drug metabolism [44].
Methodology: Integrated In Vitro-In Silico Workflow
Experimental Design and PBPK Model Construction
The integration of PBPK modeling with in vitro data follows a systematic workflow that combines experimental assays with computational simulations. A recent study investigating developmental toxicity thresholds for known reproductive toxicants exemplifies this integrated approach, employing a three-tiered PBPK modeling framework to translate in vitro concentrations from ReproTracker, Stemina DevTOX quickPredict, and developmental neurotoxicity (DNT) battery assays into human equivalent doses (HEDs) [53]. This methodology demonstrates how in vitro screening platforms for teratogenic potential can be quantitatively connected to human exposure levels through PBPK modeling, advancing the reduction of animal testing while enhancing human-relevant toxicity assessment [53].
PBPK model construction involves two fundamental components: an anatomical "backbone" containing species-specific physiological parameters independent of the drug, and a drug-specific part consisting of the individual drug's ADME properties applied to relevant processes within each tissue compartment [51]. Parameters for PBPK models are either drug-dependent (e.g., binding to blood, fraction unbound; tissue-to-plasma distribution coefficient, KPT; enzymatic activity, Vmax/Km) or drug-independent (e.g., blood flows, tissue volumes, tissue composition) [51]. The accuracy of PBPK prediction depends not only on current knowledge of animal or human physiology but also on the physiochemical and biochemical properties of the test compounds [51].
Table 1: Essential Drug-Dependent Parameters for PBPK Model Development
| Parameter Category | Specific Parameters | Commonly Used Assays/Approaches |
|---|---|---|
| Physicochemical Properties | Molecular weight, logP, pKa, compound type (base, acid, neutral) | Physicochemistry property measurement, in silico prediction |
| Solubility & Permeability | pH-dependent solubility, apparent permeability | Measured in buffer with different pH; Caco-2, MDCK assays |
| Binding & Partitioning | Plasma protein binding (fub), blood-plasma partitioning (B:P) | In vitro in human plasma and blood |
| Metabolic Parameters | Intrinsic clearance (CLint), Vmax and Km, percent enzyme contribution (fm) | Microsomes, S9, hepatocytes, or recombinant CYP systems |
| Inhibition/Induction Potential | Reversible inhibition IC50, mechanism-based inhibition (kinact, KI), induction (Jmax, EC50) | Human liver microsomes, human hepatocytes |
In Vitro Assays and Experimental Protocols
The experimental foundation for PBPK modeling relies on robust in vitro assays that provide quantitative parameters for model input. For metabolic clearance prediction, standardized protocols using human liver microsomes, S9 fractions, or hepatocytes are employed to determine intrinsic clearance (CLint) values [51]. These assays typically involve incubating test compounds with the metabolic system at relevant protein concentrations (e.g., 0.5-1 mg/mL for microsomes) for predetermined timepoints, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis to quantify parent compound depletion [51]. Reaction phenotyping studies to identify specific enzymes involved in metabolism utilize chemical inhibitors, recombinant enzymes, or correlation analyses to determine the fraction metabolized (fm) by each pathway [44].
For transporter-mediated interactions, polarized cell lines overexpressing specific transporters (e.g., MDCKII-MDR1 for P-glycoprotein) are used to assess bidirectional transport, generating parameters such as the transporter-mediated efflux ratio [51]. Plasma protein binding determinations employ equilibrium dialysis or ultrafiltration methods to measure the fraction unbound (fub) in plasma, while blood-to-plasma partitioning ratios are determined by incubating compounds in fresh blood and measuring distribution between blood cells and plasma [51]. These experimentally derived parameters provide critical input for PBPK models, enabling more accurate predictions of in vivo pharmacokinetics.
Case Study: Application to Complex Molecules
PBPK Modeling for Special Populations and DDI Predictions
PBPK modeling demonstrates particular value for complex molecules where clinical testing presents significant challenges. In populations with specific physiological characteristics—such as pediatrics, geriatrics, pregnant women, and individuals with organ impairments—PBPK models can virtualize pharmacokinetic profiles by incorporating population-specific physiological parameters [44] [52]. This approach is particularly valuable for simulating the altered drug exposure resulting from pathophysiological changes in key organs such as the gut and liver, as well as for accounting for the distinct physiological characteristics of special populations [52]. Consequently, PBPK modeling provides crucial support for designing initial dosing regimens in groups where large-scale clinical trials are challenging [52].
The quantitative prediction of drug-drug interactions (DDIs) constitutes the predominant application of PBPK modeling in regulatory submissions, representing 81.9% of all instances in recent FDA approvals [52]. A granular breakdown of DDI mechanisms reveals that enzyme-mediated interactions (primarily CYP3A4) account for the majority (53.4%), followed by transporter-mediated interactions (e.g., P-gp, 25.9%) [52]. PBPK models can dynamically and quantitatively predict the impact on substrate drug exposure by simulating the kinetics of metabolic enzyme or transporter inhibition/induction, thereby informing clinical risk management strategies for combination therapies [52].
Table 2: PBPK Model Evaluation Against Clinical Observed Data
| Application Domain | Number of Instances | Percentage of Total | Key Predictive Performance Metrics |
|---|---|---|---|
| Drug-Drug Interactions (DDI) | 95 | 81.9% | Prediction within 1.25-fold for AUC ratio; 2.0-fold for Cmax ratio |
| Enzyme-Mediated DDI | 62 | 53.4% | Geometric mean fold error (GMFE) <1.25 for AUC ratio |
| Transporter-Mediated DDI | 30 | 25.9% | Prediction within 2.0-fold of observed values |
| Organ Impairment Dosing | 8 | 7.0% | Prediction within 25% of observed exposure parameters |
| Hepatic Impairment | 5 | 4.3% | Prediction of exposure changes within 30% of observed |
| Renal Impairment | 3 | 2.6% | Prediction of clearance changes within 25% of observed |
Incorporating Genetic Polymorphisms and Ethnic Factors
PBPK models can effectively incorporate genetic variations affecting drug metabolism, particularly polymorphisms in cytochrome P450 enzymes such as CYP2D6, CYP2C9, and CYP2C19 [44]. These genetic differences can result in ultrarapid, rapid, intermediate, or poor metabolism phenotypes, significantly impacting drug exposure and response [44]. By integrating population-specific allele frequency data and the associated changes in enzyme activity, PBPK models can simulate the range of expected exposures in ethnically diverse populations, informing personalized dosing strategies [44].
Ethnic influences in PBPK modeling extend beyond genetic polymorphisms to include physiological variations such as differences in enzyme abundances and liver volume [44]. Commercial PBPK platforms now include distinct metabolic profiles for various ethnic populations, including Caucasian, Japanese, and Chinese populations [44]. Additionally, lifestyle factors more prevalent in certain populations—such as coffee consumption or smoking, which increase CYP1A2 activity—can be incorporated to improve the accuracy of pharmacokinetic predictions across different ethnic groups [44].
Validation and Regulatory Considerations
Model Credibility and Verification
As PBPK models are increasingly included in regulatory submissions, establishing model credibility through rigorous verification and validation processes has become essential [54]. The ASME VV-40-2018 standard, "Assessing Credibility of Computational Modeling through Verification and Validation: Application to Medical Devices," introduces a risk-informed credibility assessment framework that begins with defining the Context of Use (COU)—the specific role and scope of the model in addressing the question of interest [54]. With a well-defined COU, model risk can be identified based on the combination of model influence (the contribution of the model to the decision) and decision consequence (the impact of an incorrect decision) [54].
Model verification ensures that the computational model has been implemented correctly and operates as intended, addressing numerical accuracy, software quality, and calculation verification [54]. This process includes confirming that the governing equations have been solved correctly, input parameters have been appropriately incorporated, and the model produces stable, convergent solutions across the intended domain of application [54]. For PBPK models, verification typically involves comparing simulated concentrations against analytical solutions for simplified cases, ensuring mass balance is maintained throughout simulations, and verifying that parameter sensitivities align with physiological expectations [54].
Experimental Validation and Regulatory Acceptance
Model validation provides evidence that the computational model accurately represents the real-world system for the specific Context of Use [54]. For PBPK models, this involves comparing simulated pharmacokinetic parameters (e.g., AUC, Cmax, clearance) against observed clinical data not used in model development [54] [52]. Successful validation typically requires predictions to fall within predefined acceptance criteria, often expressed as a percentage of predictions within two-fold of observed values or geometric mean fold error (GMFE) thresholds [52].
Regulatory agencies have established formal guidelines for PBPK model submissions, reflecting the growing acceptance of this technology in drug development. In September 2018, the U.S. FDA issued the industry guidance "Physiologically Based Pharmacokinetic Analyses—Format and Content," formally recognizing the regulatory role of PBPK in drug-drug interaction assessments [52]. Similarly, in December 2018, the European Medicines Agency (EMA) published its first PBPK-specific guideline, "Guideline on the Reporting of Physiologically Based Pharmacokinetic (PBPK) Modelling and Simulation," marking the institutional recognition of this technology within the international regulatory framework [52].
The Scientist's Toolkit: Essential Research Reagents and Platforms
Table 3: Key Research Reagent Solutions for PBPK Modeling
| Tool Category | Specific Tools/Reagents | Function in PBPK Workflow |
|---|---|---|
| In Vitro Metabolic Systems | Human liver microsomes, hepatocytes, recombinant CYP enzymes, S9 fractions | Determination of intrinsic clearance (CLint), reaction phenotyping, inhibition parameters |
| Transporter Assay Systems | Polarized cell lines (MDCK, Caco-2), vesicular transport assays, transfected cell lines | Assessment of transporter-mediated uptake/efflux, DDI potential |
| Protein Binding Assays | Equilibrium dialysis devices, ultrafiltration systems, rapid equilibrium dialysis (RED) | Measurement of fraction unbound in plasma (fub) and tissue homogenates |
| PBPK Software Platforms | Simcyp, GastroPlus, PK-Sim, Cloe PK | Integrated platforms for PBPK model development, simulation, and validation |
| Bioanalytical Instruments | LC-MS/MS systems, high-resolution mass spectrometers, automated sample preparation | Quantification of drug concentrations in in vitro assays and biological matrices |
The integration of PBPK modeling with in vitro data represents a transformative approach in drug development, enabling more predictive assessment of complex pharmacokinetic scenarios while reducing reliance on animal testing and extensive clinical trials [53]. This case study demonstrates how mechanistic PBPK models, parameterized with robust in vitro data, can successfully predict human pharmacokinetics, inform dosing recommendations for special populations, and assess drug interaction potential [44] [52]. The growing regulatory acceptance of PBPK modeling, evidenced by its inclusion in over 26% of recent FDA submissions, underscores its value in modern drug development [52].
Looking forward, the integration of PBPK modeling with artificial intelligence (AI) and multi-omics data will unprecedentedly enhance predictive accuracy, thereby providing critical and actionable insights for decision-making in precision medicine and global regulatory strategies [52]. Emerging approaches such as federated learning show promise for collaboratively training ADMET models across distributed proprietary datasets without compromising data confidentiality, systematically extending the model's effective domain beyond what can be achieved with isolated internal datasets [9]. Additionally, initiatives like OpenADMET aim to generate high-quality, consistent experimental data specifically for ADMET model development, addressing current limitations associated with heterogeneous literature data [55]. These advancements, combined with rigorous model validation frameworks and standardized experimental protocols, will further establish PBPK modeling as an indispensable tool for optimizing the efficiency and reliability of drug development.
Visual Workflows
PBPK Modeling Workflow
Model Validation Framework
The accurate prediction of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties remains a fundamental challenge in modern drug discovery, with approximately 40-45% of clinical attrition still attributed to ADMET liabilities [9]. This challenge is particularly acute for natural products and new therapeutic modalities, which often exhibit unique structural complexity and fall outside the chemical space of conventional drug-like compounds [1]. The validation of computational ADMET predictions against experimental data has therefore become a critical frontier, bridging the promise of in silico methods with the practical demands of drug development.
Natural compounds present specific validation challenges due to their structural diversity, complex stereochemistry, and frequently limited availability for experimental testing [1]. Meanwhile, emerging modalities push the boundaries of traditional quantitative structure-activity relationship (QSAR) models. This comparison guide examines current methodologies, benchmarking frameworks, and experimental protocols that are advancing the validation of ADMET predictions, enabling researchers to select appropriate strategies for their specific discovery context.
Methodological Frameworks for ADMET Validation
Systematic Benchmarking Approaches
Rigorous benchmarking provides the foundation for meaningful comparison of ADMET prediction methods. The ADMET Benchmark Group has established standardized frameworks that curate diverse datasets from sources like ChEMBL and TDC (Therapeutics Data Commons), employing scaffold-based, temporal, and out-of-distribution splits to ensure robust evaluation [49]. These benchmarks encompass numerous ADMET endpoints—including lipophilicity, solubility, CYP inhibition, membrane permeability, and toxicity markers—enabling comprehensive assessment of predictive accuracy across the entire pharmacokinetic and safety spectrum [49].
These benchmarking initiatives systematically evaluate a wide spectrum of modeling approaches, from classical machine learning to advanced deep learning architectures. The validation typically employs multiple metrics tailored to the specific prediction task: Mean Absolute Error (MAE) and Root Mean Squared Error (RMSE) for regression endpoints (e.g., logD, solubility), and Area Under the ROC Curve (AUROC), Area Under the Precision-Recall Curve (AUPRC), and Matthews Correlation Coefficient (MCC) for classification tasks (e.g., toxicity flags, CYP inhibition) [49]. This multi-faceted evaluation provides a more complete picture of model performance than any single metric could offer.
Integrated Validation Protocols
Beyond standard benchmarking, advanced validation protocols incorporate statistical hypothesis testing alongside cross-validation to provide greater confidence in model selection, particularly important in noisy ADMET prediction tasks [14]. This approach combines k-fold cross-validation with statistical tests like paired t-tests to determine whether performance differences between models are statistically significant rather than merely incidental [14].
The most robust validation frameworks also include "practical scenario" testing, where models trained on data from one source (e.g., public databases) are evaluated against test sets from different sources (e.g., proprietary corporate data) [14]. This assesses real-world generalizability, mimicking the actual application of models across organizational boundaries and experimental protocols. For natural products specifically, validation often includes scaffold-based splits that separate structurally distinct compounds between training and test sets, ensuring models can generalize to novel chemotypes rather than merely memorizing similar structures [14] [1].
Comparative Performance of ADMET Prediction Methods
Machine Learning Model Performance
Table 1: Performance comparison of major ADMET prediction approaches across benchmark datasets
| Model Class | Feature Modalities | Key Strengths | Validation Performance | Best Applications |
|---|---|---|---|---|
| Random Forest / GBDT | ECFP, RDKit descriptors, Mordred | High interpretability, robust on small data | State-of-the-art on several TDC benchmarks [49] | Early screening, limited data contexts |
| Graph Neural Networks (GAT, MPNN) | Molecular graph, learned embeddings | Structure-aware, no feature engineering needed | Superior OOD generalization (GAP: AUCID - AUCOOD = 2-5%) [49] | Novel scaffold prediction, natural products |
| Multimodal Models (MolIG) | Graph + molecular image | Multiple representation learning | Outperforms single-modal baselines on permeability [49] | Complex property prediction |
| Foundation Models | SMILES sequence, quantum properties | Transfer learning, reduced data needs | Top-1 on diverse benchmarks after fine-tuning [49] | Data-scarce endpoints, new modalities |
| AutoML Frameworks (Auto-ADMET) | Dynamic feature selection | Adaptive to specific datasets | Personalized performance, best on several ADMET endpoints [49] | Automated pipelines, non-expert users |
The benchmarking studies reveal that no single algorithm universally dominates all ADMET prediction tasks. Instead, optimal model selection depends on factors including data volume, chemical space, and the specific ADMET property being predicted. Ensemble methods like random forests and gradient-boosted decision trees (GBDT) remain remarkably competitive, particularly with systematic feature combination and optimization [14] [49]. These classical approaches demonstrate particular strength in contexts with limited training data, offering robust performance without extensive hyperparameter tuning.
Graph neural networks (GNNs), especially graph attention networks (GATs) and message passing neural networks (MPNNs), have shown excellent generalization to out-of-distribution compounds, a critical capability for predicting ADMET properties of natural products with novel scaffolds [49]. The end-to-end representation learning in GNNs eliminates the need for manual feature engineering, allowing the models to discover relevant structural motifs directly from atomic connectivity data [14] [33]. This advantage makes GNNs particularly valuable for natural products, which often contain structural features poorly captured by conventional molecular descriptors.
Impact of Representation and Feature Selection
The choice of molecular representation significantly influences prediction accuracy, sometimes exceeding the impact of the model architecture itself. Benchmarking studies have systematically evaluated diverse feature representations including fingerprints (ECFP, FCFP), molecular descriptors (RDKit, Mordred), and deep-learned embeddings [14]. The conventional practice of concatenating multiple representations without systematic reasoning often yields good benchmark performance but may limit interpretability and generalizability [14].
Table 2: Feature representation performance in ADMET prediction
| Representation Type | Examples | Advantages | Validation Performance | Limitations |
|---|---|---|---|---|
| Fingerprints | ECFP4, FCFP4, Avalon | Computational efficiency, interpretability | Strong in classical models [14] | Limited structural insight |
| Molecular Descriptors | RDKit descriptors, Mordred | Physicochemically meaningful | Good for solubility, logP [14] | Manual engineering required |
| Deep-learned Representations | MPNN embeddings, SMILES embeddings | Automatic feature learning | Superior on complex endpoints [14] [33] | Black box, data hungry |
| Multimodal Representations | Graph + image, sequence + structure | Complementary information | Enhanced robustness [49] | Increased complexity |
| Quantum Chemical Features | DFT-calculated properties | Fundamental molecular characteristics | Strong for metabolism [1] [11] | Computational cost |
Recent research indicates that structured approaches to feature selection—iteratively evaluating representation combinations and selecting optimal sets for specific datasets—can yield more interpretable and generalizable models than simply concatenating all available features [14]. For natural products, quantum chemical descriptors derived from density functional theory (DFT) calculations have shown particular value for predicting metabolic properties, capturing electronic characteristics that influence interactions with metabolic enzymes like cytochromes P450 [1].
Experimental Protocols for Validation
Data Curation and Cleaning Standards
Robust validation begins with rigorous data curation. Current best practices include comprehensive cleaning procedures to address common issues in public ADMET datasets: inconsistent SMILES representations, duplicate measurements with varying values, fragmented structures, and contradictory labels for the same compound across different sources [14]. The protocol typically includes:
- Standardization of representation: Using tools like the standardisation tool by Atkinson et al. to generate consistent SMILES strings, with modifications to account for organic elements like boron and silicon [14].
- Salt removal and parent compound extraction: Particularly important for solubility datasets where salt forms can obscure the intrinsic properties of the parent compound [14].
- Tautomer standardization: Ensuring consistent representation of tautomeric forms [14].
- Duplicate handling: Keeping the first entry if target values are consistent, or removing the entire group if inconsistencies exceed a defined threshold (e.g., beyond 20% of inter-quartile range for regression tasks) [14].
- Visual inspection: Using tools like DataWarrior for final data quality assessment, especially important with smaller datasets [14].
For natural products, additional curation considerations include handling of complex stereochemistry, representative sampling of diverse scaffolds, and addressing the higher prevalence of reactive functional groups that may represent pan-assay interference compounds (PAINS) [1].
Validation Workflows for Natural Products
The complex structure and unique challenges of natural products necessitate specialized validation workflows. The following diagram illustrates an integrated approach that combines multiple computational and experimental methods:
Integrated Validation Workflow for Natural Products
This workflow emphasizes the iterative nature of validation, where discrepancies between computational predictions and experimental results feed back into model refinement, creating a continuous improvement cycle [1] [56] [57]. For natural products, this process often requires special consideration of their distinct chemical space, which includes more oxygen atoms, more chiral centers, and larger molecular size compared to synthetic compounds [1].
Statistical Validation Methods
Robust statistical validation goes beyond simple train-test splits to include:
- Scaffold-based cross-validation: Ensuring that structurally dissimilar compounds are separated between training and test sets, providing a more realistic assessment of model generalizability to novel chemotypes [14] [56].
- Temporal splits: Mimicking real-world application scenarios by training on older data and testing on newer compounds, assessing temporal generalizability [49].
- Statistical hypothesis testing: Using paired t-tests or similar methods to determine whether performance differences between models are statistically significant rather than random variations [14].
- Uncertainty quantification: Especially important for decision-making in drug discovery, with Gaussian Process models and Bayesian neural networks providing confidence estimates alongside predictions [14].
These rigorous statistical approaches help distinguish genuinely improved modeling strategies from those that merely capitalize on chance variations in specific datasets.
Visualization of Key Methodological Relationships
ADMET Model Comparison Framework
The landscape of ADMET prediction methods can be understood through their relationships across key dimensions relevant to validation. The following diagram maps major approaches according to their methodological characteristics and validation strengths:
ADMET Model Comparison Framework
This framework highlights how different approaches offer complementary strengths, suggesting that ensemble methods or hybrid pipelines may often provide optimal performance across diverse validation scenarios [14] [33] [49].
Research Reagent Solutions for ADMET Validation
Computational Tools and Platforms
Table 3: Essential research reagents and computational tools for ADMET validation
| Tool Category | Specific Tools | Primary Function | Application in Validation |
|---|---|---|---|
| Benchmark Platforms | TDC [14], ADMEOOD [49] | Standardized datasets and evaluation | Method comparison, baseline establishment |
| Molecular Representation | RDKit [14], Mordred, PaDEL | Fingerprint and descriptor calculation | Feature engineering, representation comparison |
| Machine Learning | Scikit-learn, XGBoost, CatBoost [14] | Classical ML implementation | Baseline models, ensemble methods |
| Deep Learning | Chemprop [14], DeepChem, PyTorch Geometric | Graph neural network implementation | End-to-end learning, complex relationship modeling |
| ADMET Prediction | SwissADME [56], admetSAR [57], PreADMET [58] | Property-specific prediction | Result verification, multi-tool consensus |
| Simulation & Dynamics | GROMACS, Desmond [56] [57] | Molecular dynamics simulation | Mechanism understanding, binding stability |
| Quantum Chemistry | Gaussian [59], ORCA | Electronic structure calculation | Metabolic prediction, reactivity assessment |
These tools collectively enable comprehensive validation pipelines, from initial screening to mechanistic understanding. The integration of multiple tools provides cross-validation and enhances confidence in predictions, particularly important for natural products where single-model predictions may be less reliable [1] [56] [57].
The validation of ADMET predictions for natural products and new modalities remains a challenging but rapidly advancing field. Current benchmarking indicates that while classical machine learning methods retain strong performance on many endpoints, graph neural networks and multimodal approaches offer superior generalizability to novel chemical spaces—a critical capability for natural product applications [14] [49]. The most robust validation strategies combine rigorous statistical assessment with practical scenario testing, ensuring models deliver not just theoretical accuracy but practical utility in real drug discovery settings.
Future directions point toward increased use of federated learning to expand chemical diversity without compromising data privacy [9], more sophisticated uncertainty quantification to guide experimental prioritization [14], and enhanced focus on out-of-distribution robustness through benchmarks like ADMEOOD and DrugOOD [49]. As these methodologies mature, integrated validation frameworks will become increasingly essential for selecting the optimal ADMET prediction strategy for specific discovery contexts, particularly for challenging compound classes like natural products that push the boundaries of conventional chemical space.
Overcoming Key Challenges and Optimizing Predictive Performance
The application of artificial intelligence (AI) in drug discovery, particularly for predicting absorption, distribution, metabolism, excretion, and toxicity (ADMET) properties, has transformed the pharmaceutical research landscape. However, the most advanced AI systems remain 'black boxes' whose inner workings even their developers cannot fully understand [60]. This opacity creates significant challenges for researchers, scientists, and drug development professionals who require not just predictions but understandable reasoning behind them to make critical decisions in the drug development pipeline. The lack of transparency can lead to decreased trust, reduced adoption, and increased regulatory scrutiny [61], particularly concerning when AI-driven predictions are validated against in vitro data.
The bipartisan National Security Commission on AI has cautioned that AI systems perceived as unreliable or unpredictable will 'stall out': leaders will not adopt them, operators will mistrust them, and Congress will not fund them [60]. In high-stakes fields like drug development, where approximately 30% of drug candidates fail due to inadequate pharmacokinetic properties [62] and 40-45% of clinical attrition continues to be attributed to ADMET liabilities [9], explainability transitions from a technical nicety to a practical necessity. This guide explores strategies for improving AI model interpretability, comparing various approaches within the critical context of validating in silico ADMET predictions with in vitro experimental data.
The Black Box Problem & Explainable AI (XAI) Fundamentals
The Transparency Crisis in Modern AI
The increasing complexity of AI models, particularly deep learning, has led to a significant challenge: the black box problem. This refers to the lack of transparency and interpretability in AI decision-making processes, making it difficult to understand how models arrive at their predictions or recommendations [63]. As AI becomes more pervasive in critical applications like healthcare and drug discovery, the risks associated with black box decision-making become more pronounced.
In pharmaceutical research, AI models are being used to predict ADMET properties and diagnose diseases. However, if these models are not transparent, it can be difficult to understand why a particular prediction was made. This lack of transparency can lead to mistrust among researchers and clinicians, ultimately undermining the adoption of AI in healthcare. According to a study, explaining AI models in medical imaging can increase the trust of clinicians in AI-driven diagnoses by up to 30% [63].
The black box problem is not limited to these sectors; it is a broader issue that affects many industries. The lack of transparency in AI decision-making processes can lead to:
- Unintended biases: AI models can perpetuate existing biases and discriminate against certain groups, which can have significant social and economic consequences [63].
- Errors and inaccuracies: AI models can make mistakes, which can have significant consequences in critical applications [63].
- Lack of trust: The lack of transparency in AI decision-making processes can lead to mistrust among stakeholders, ultimately undermining the adoption of AI [63].
To address the black box problem, there is a growing need for explainable AI (XAI) techniques and tools. As of 2025, the XAI market size is projected to be $9.77 billion, up from $8.1 billion in 2024, with a compound annual growth rate (CAGR) of 20.6% [63]. Companies like IBM and Google are investing heavily in XAI research and development, and experts like Dr. David Gunning, Program Manager at DARPA, emphasize that "explainability is not just a nice-to-have, it's a must-have for building trust in AI systems" [63]. By providing transparent and interpretable models, XAI can help mitigate the risks associated with black box decision-making and increase trust in AI systems.
Key Concepts: Transparency vs. Interpretability
When it comes to explainable AI, two concepts are often thrown around: transparency and interpretability. While they're related, they're not interchangeable terms. Think of transparency like looking at a car's engine – you can see all the parts and understand how they work together. Interpretability, on the other hand, is like understanding why the car's navigation system took a specific route – you want to know the reasoning behind the decision [63].
Transparency refers to the ability to understand how a model works, including its architecture, algorithms, and data used to train it. It's about opening up the "black box" and shedding light on the inner workings of the AI system. For instance, IBM's AI Explainability 360 toolkit provides a suite of algorithms and techniques to help explain AI models, enhancing transparency and trust in AI decision-making processes [63].
Interpretability, however, is about understanding why a model makes specific decisions. It's about understanding the relationships between the input data, the model's parameters, and the output predictions. In other words, interpretability helps you understand the "why" behind the model's predictions. For example, in healthcare, interpretability can help researchers understand why a particular drug candidate was predicted to have toxic effects [63].
The Business and Regulatory Case for XAI
The business case for Explainable AI (XAI) in 2025 is stronger than ever, with the market projected to reach $9.77 billion, up from $8.1 billion in 2024, with a compound annual growth rate (CAGR) of 20.6% [63]. This growth is driven by the increasing need for transparency and accountability in AI systems, particularly in sectors such as healthcare, education, and finance. For businesses, XAI offers numerous tangible benefits, including regulatory compliance, improved model debugging, enhanced user trust, and ethical considerations [63].
One of the primary advantages of XAI is its ability to facilitate regulatory compliance. With the implementation of industry standards and regulatory requirements, such as GDPR and healthcare compliance standards, companies can ensure that their AI systems are transparent and accountable. For instance, IBM's AI Explainability 360 toolkit provides a suite of algorithms and techniques to help explain AI models, enhancing transparency and trust in AI decision-making processes [63]. This is particularly important in sectors like healthcare and finance, where interpretability and accountability are crucial.
XAI also enables improved model debugging, allowing developers to identify and address errors in AI models more efficiently. By providing insights into how AI models make decisions, XAI facilitates the identification of biases and flaws, ultimately leading to more accurate and reliable AI systems. A study using XAI techniques found that explaining AI models in medical imaging can increase the trust of clinicians in AI-driven diagnoses by up to 30% [63].
Enhanced user trust is another significant benefit of XAI. By providing transparency into AI decision-making processes, businesses can build trust with their customers and stakeholders. For example, Google's Model Interpretability platform allows developers to understand how their AI models are making predictions, enabling them to make more informed decisions and build more trustworthy AI systems [63].
Furthermore, XAI has important ethical considerations. As AI becomes more pervasive, the need for transparent and interpretable models will only increase. Dr. David Gunning, Program Manager at DARPA, notes that "Explainability is not just a nice-to-have, it's a must-have for building trust in AI systems" [63]. By prioritizing XAI, businesses can ensure that their AI systems are fair, accountable, and transparent, ultimately leading to more responsible and ethical AI adoption.
Comparative Analysis of AI Interpretability Approaches
Technical Approaches for Explainable AI
Local vs. Global Explanations
When it comes to understanding AI explainability techniques, it's essential to grasp the difference between local explanations and global explanations. Local explanations focus on providing insights into individual predictions made by a model, whereas global explanations aim to understand the model as a whole, including its behavior, biases, and decision-making processes [61].
Local explanations are particularly valuable in situations where a specific prediction or outcome needs to be understood, such as in medical diagnosis or credit risk assessment. For instance, IBM's AI Explainability 360 tool provides local explanations by generating feature importance scores for individual predictions, allowing users to comprehend how the model arrived at a specific decision. In a Google research study, local explanations were used to analyze the performance of a machine learning model in a medical diagnosis task, revealing that the model was relying heavily on a specific feature that was not relevant to the diagnosis [61].
On the other hand, global explanations are crucial for understanding the overall behavior of a model, including its strengths, weaknesses, and potential biases. This is particularly important in high-stakes applications, such as autonomous vehicles or financial modeling. SuperAGI's Transparency Suite, for example, provides global explanations by analyzing the model's behavior across a large dataset, identifying patterns and biases that may not be apparent from individual predictions. A study by McKinsey found that global explanations can help reduce model bias by up to 30%, resulting in more accurate and reliable predictions [61].
Some key differences between local and global explanations include:
- Scope: Local explanations focus on individual predictions, while global explanations examine the model as a whole.
- Purpose: Local explanations aim to understand a specific outcome, while global explanations seek to comprehend the model's behavior and decision-making processes.
- Methodology: Local explanations often rely on feature importance scores or partial dependence plots, while global explanations use techniques such as model interpretability methods or sensitivity analysis [61].
In terms of current trends, the market for AI explainability tools is expected to grow by 25% in the next year, driven by increasing demand for transparent and accountable AI systems. According to a survey by Gartner, 75% of organizations consider explainability to be a critical factor in their AI adoption decisions. As the field of explainable AI continues to evolve, it's essential to understand the differences between local and global explanations and how they can be used to build more transparent, trustworthy, and effective AI systems [61].
By leveraging both local and global explanations, organizations can gain a deeper understanding of their AI models and make more informed decisions. For example, a company like Google can use local explanations to analyze the performance of its search algorithm, while also using global explanations to understand how the algorithm is behaving across different regions and user demographics. By combining these insights, Google can refine its algorithm to provide more accurate and relevant search results, while also ensuring that it is fair and unbiased [61].
Technical vs. User-Friendly Explanations
When it comes to AI explainability, the target audience plays a significant role in determining the type of explanation required. Different stakeholders, such as technical ML engineers, business stakeholders, and end users, have varying levels of expertise and needs. As a result, explainability tools differ in their approach to cater to these diverse audiences. For instance, tools like IBM AI Explainability 360 and LIME provide technical explanations, focusing on model interpretability and feature importance [61].
Technical explanations are designed for ML engineers and data scientists who have a deep understanding of AI models and their inner workings. These explanations provide detailed insights into the model's architecture, parameters, and decision-making processes. They often include feature importance scores, partial dependence plots, and other technical metrics that help engineers debug and improve the model's performance.
On the other hand, user-friendly explanations are tailored for business stakeholders and end users who may not have a technical background. These explanations focus on providing clear, concise, and actionable insights into the model's decisions, without overwhelming the user with technical details. For example, a user-friendly explanation might highlight the key factors that influenced a particular prediction, such as "the compound was predicted to be toxic due to its high molecular weight and low solubility."
The choice between technical and user-friendly explanations depends on the audience and the purpose of the explanation. Technical explanations are essential for model development and debugging, while user-friendly explanations are crucial for building trust and facilitating decision-making among non-technical stakeholders.
Comparative Analysis of XAI Tools and Platforms
The growing demand for explainable AI has led to the development of numerous tools and platforms designed to enhance transparency and interpretability. These tools vary in their approach, capabilities, and target audiences. The following table provides a comparative analysis of some of the top XAI tools available in 2025:
Table 1: Comparative Analysis of XAI Tools and Platforms
| Tool/Platform | Developer | Primary Approach | Key Features | Best For |
|---|---|---|---|---|
| AI Explainability 360 | IBM | Model-agnostic explanations | Comprehensive algorithm collection, local and global explanations | Technical users, model debugging |
| Model Interpretability Platform | Integrated with Google Cloud AI | What-if tool, feature attribution, fairness indicators | Google Cloud users, enterprise applications | |
| SHAP (SHapley Additive exPlanations) | Open-source | Game theory-based | Unified measure of feature importance, local explanations | Academic research, technical users |
| LIME (Local Interpretable Model-agnostic Explanations) | Open-source | Local surrogate models | Explains individual predictions, model-agnostic | Technical users, model validation |
| DALEX (Descriptive Automated Learning EXplanation) | Open-source | Model-agnostic explanations | Model performance, variable importance, residual diagnostics | R users, model auditing |
These tools offer a range of capabilities, from model-agnostic explanations to integrated platforms that provide end-to-end interpretability solutions. The choice of tool depends on the specific needs of the user, the type of model being explained, and the audience for the explanations.
Interpretability in Practice: ADMET Prediction Case Studies
Benchmarking Machine Learning for ADMET Predictions
Accurate prediction of ADMET properties remains a fundamental challenge in drug discovery. Despite the progress of graph-based deep learning and foundation models, even the most advanced approaches continue to be constrained by the data on which they are trained. Experimental assays are heterogeneous and often low-throughput, while available datasets capture only limited sections of chemical and assay space. As a result, model performance typically degrades when predictions are made for novel scaffolds or compounds outside the distribution of training data [9].
Recent benchmarking initiatives such as the Polaris ADMET Challenge have made this issue explicit. Multi-task architectures trained on broader and better-curated data consistently outperformed single-task or non-ADMET pre-trained models, achieving up to 40–60% reductions in prediction error across endpoints including human and mouse liver microsomal clearance, solubility (KSOL), and permeability (MDR1-MDCKII). These results highlight that data diversity and representativeness, rather than model architecture alone, are the dominant factors driving predictive accuracy and generalization [9].
A recent study focusing on predicting ADMET properties addressed the key challenges of ML models trained using ligand-based representations. The researchers proposed a structured approach to data feature selection, taking a step beyond the conventional practice of combining different representations without systematic reasoning. Additionally, they enhanced model evaluation methods by integrating cross-validation with statistical hypothesis testing, adding a layer of reliability to the model assessments. The final evaluations included a practical scenario, where models trained on one source of data were evaluated on a different one. This approach aims to bolster the reliability of ADMET predictions, providing more dependable and informative model evaluations [14].
The study conducted experiments to enlighten the following research questions:
- Which types of algorithms and compound representations are generally suitable for ligand-based machine learning in the ADMET domain?
- Can cross-validation hypothesis testing serve as a more robust model comparison than a hold-out test set in the ADMET domain?
- How important are various forms of model optimization in a practical scenario?
- What is the impact on the model performance when available external data of the same property is used in combination with internal data? [14]
Experimental Protocols for Robust ADMET Model Validation
The benchmarking study employed rigorous experimental protocols to ensure robust and reliable model validation. The experiments were carried out sequentially, achieving the following:
- A model architecture is chosen to use as a baseline as well as optimize in further experiments;
- Features are combined iteratively until the best-performing combinations are identified;
- Hyperparameters of the chosen model architecture are tuned in a dataset-specific manner;
- Cross-validation hypothesis testing is done in order to assess the statistical significance of the optimization steps;
- Test set performance is evaluated, assessing the impact of the previous optimization steps, as well as the contrast between the hypothesis test outcomes and test set changes;
- The optimized models are evaluated in a practical scenario, where models trained on one data source are evaluated on a test set from a different source, for the same property and;
- Finally, the optimized model is trained on a combination of data from two different sources, to mimic the scenario when external data is combined with increasing amounts of internal data [14].
The machine learning algorithms included in the study ranged from classical models to more recent neural networks. Included is Support Vector Machines (SVM), tree-based methods comprising Random Forests (RF) and gradient boosting frameworks LightGBM and CatBoost, as well as Message Passing Neural Networks (MPNN) as implemented by Chemprop [14].
Various descriptors, fingerprints, and embeddings were used on their own or in combination. The following descriptors and fingerprints were implemented using the RDKit cheminformatics toolkit: RDKit descriptors (rdkit_desc), Morgan fingerprints (morgan), and others. These features were combined iteratively to identify the best-performing combinations for each dataset [14].
The following workflow diagram illustrates the experimental protocol for robust ADMET model validation:
Diagram 1: Experimental Protocol for Robust ADMET Model Validation. This workflow outlines the systematic approach for developing and validating interpretable ADMET prediction models, emphasizing statistical rigor and practical applicability.
Federated Learning for Enhanced ADMET Prediction
Because each organization's assays describe only a small fraction of the relevant chemical space, isolated modeling efforts remain inherently limited. Federated learning provides a method to overcome this limitation by enabling model training across distributed proprietary datasets without centralizing sensitive data. Cross-pharma research has already provided a consistent picture of the advantages of this approach [9]:
- Federation alters the geometry of chemical space a model can learn from, improving coverage and reducing discontinuities in the learned representation
- Federated models systematically outperform local baselines, and performance improvements scale with the number and diversity of participants
- Applicability domains expand, with models demonstrating increased robustness when predicting across unseen scaffolds and assay modalities
- Benefits persist across heterogeneous data, as all contributors receive superior models even when assay protocols, compound libraries, or endpoint coverage differ substantially
- Multi-task settings yield the largest gains, particularly for pharmacokinetic and safety endpoints where overlapping signals amplify one another
Together, these findings suggest that federation systematically extends the model's effective domain, an effect that cannot be achieved by expanding isolated internal datasets [9].
At Apheris, every ADMET model follows recommended practices to deliver results partners can truly rely on. For pre-trained models, they carefully validate datasets (performing sanity and assay consistency checks) with normalization. Data is then sliced by scaffold, assay, and activity cliffs, ensuring they grasp modelability before training begins. With this solid foundation, they move to modeling, where ADMET models are trained and evaluated using scaffold-based cross-validation runs across multiple seeds and folds, evaluating a full distribution of results rather than a single score. Finally, the appropriate statistical tests are applied to those distributions to separate real gains from random noise [9].
In Silico ADME Methods for Natural Compounds
The pharmaceutical industry faces significant challenges when promising drug candidates fail during development due to suboptimal ADME properties or toxicity concerns. Natural compounds are subject to the same pharmacokinetic considerations. In silico approaches offer a compelling advantage—they eliminate the need for physical samples and laboratory facilities, while providing rapid and cost-effective alternatives to expensive and time-consuming experimental testing. Computational methods can often effectively address common challenges associated with natural compounds, such as chemical instability and poor solubility [1].
Through a review of the relevant scientific literature, we present a comprehensive analysis of in silico methods and tools used for ADME prediction, specifically examining their application to natural compounds. Whereas we focus on identifying the predominant computational approaches applicable to natural compounds, these tools were developed for conventional drug discovery and are of general use. We examine an array of computational approaches for evaluating natural compounds, including fundamental methods like quantum mechanics calculations, molecular docking, and pharmacophore modeling, as well as more complex techniques such as QSAR analysis, molecular dynamics simulations, and PBPK modeling [1].
Many of the challenges applicable in the understanding of the pharmacological or biological properties of natural compounds are also relevant when exploring their ADME properties. For instance, often the available quantities of natural products are limited, and while numerous plant-derived natural products have been isolated and characterized, the amounts available are frequently insufficient for comprehensive ADME testing [1]. Using in silico methods from this point of view has a great advantage as they require no physical sample (not even picograms are necessary once the structural formula is available) or laboratory infrastructure. In addition, the experimental assessment of the ADME properties of a substance is costly and time consuming, whereas the use of in silico tools is usually very cheap [1].
Key Research Reagent Solutions for ADMET Research
Table 2: Essential Research Reagents and Computational Resources for ADMET Prediction
| Category | Tool/Resource | Specific Application | Key Function | Interpretability Features |
|---|---|---|---|---|
| Cheminformatics Tools | RDKit | Compound representation | Generates molecular descriptors and fingerprints | Provides transparent feature engineering for model explanations |
| Deep Learning Frameworks | Chemprop | Message Passing Neural Networks | Specialized for molecular property prediction | Offers inherent interpretability through message passing |
| Explainable AI Toolkits | IBM AI Explainability 360 | Model-agnostic explanations | Comprehensive algorithm collection for XAI | Provides local and global explanation capabilities |
| Federated Learning Platforms | Apheris Federated ADMET Network | Cross-institutional model training | Enables collaborative training without data sharing | Enhances model generalizability and applicability domain |
| In Vitro Assay Systems | MDR1-MDCKII | Permeability assessment | Experimental measurement of drug permeability | Provides ground truth for model validation |
| In Vitro Assay Systems | Human liver microsomes | Metabolic stability testing | Experimental measurement of metabolic clearance | Enables model validation against experimental data |
Visualization of Federated Learning Workflow for ADMET
Federated learning represents a paradigm shift in how AI models for ADMET prediction can be developed across multiple institutions without sharing proprietary data. The following diagram illustrates this collaborative approach:
Diagram 2: Federated Learning Workflow for Collaborative ADMET Model Development. This approach enables multiple institutions to improve model performance while preserving data privacy and intellectual property.
The field of AI interpretability is rapidly evolving, with significant implications for drug discovery and ADMET prediction. As AI systems continue to advance toward autonomous decision-making—with minimal human oversight—AI interpretability will become not only a matter of compliance but a fundamental requirement for deploying increasingly complex and independent AI systems [64]. Organizations that proactively address this challenge, by prioritizing interpretable models and transparent processes, will be better positioned to leverage the transformative potential of AI.
The future of interpretable AI in drug discovery will likely involve several key developments. First, federated learning approaches will become more widespread, enabling collaborative model development while preserving data privacy and intellectual property. Second, advanced explanation techniques will continue to emerge, providing more nuanced and actionable insights into model behavior. Third, regulatory frameworks will increasingly require demonstrable interpretability for AI systems used in critical applications like drug development.
For researchers, scientists, and drug development professionals, the imperative is clear: embracing explainable AI is not just about building trust or meeting regulatory requirements—it's about enhancing the scientific process itself. By understanding why models make certain predictions, researchers can generate new hypotheses, identify potential pitfalls, and ultimately accelerate the development of safer, more effective therapeutics. The integration of robust interpretability techniques with rigorous experimental validation represents the most promising path forward for realizing the full potential of AI in drug discovery.
The evaluation of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties remains a critical bottleneck in drug discovery, contributing significantly to the high attrition rate of drug candidates [65]. Traditional experimental approaches for ADMET assessment, while reliable, are resource-intensive, time-consuming, and often struggle with accurately predicting human in vivo outcomes due to interspecies differences and data variability [33]. These challenges are particularly pronounced when translating results from animal models to humans, as metabolic differences between species can mask human-relevant toxicities and distort predictions [46].
The pharmaceutical industry has significantly changed its strategy in recent decades, increasingly performing extensive ADMET screening earlier in the drug discovery process to identify and eliminate problematic compounds before they enter costly development phases [1]. This shift has accelerated the adoption of in silico methods, which eliminate the need for physical samples and laboratory facilities while providing rapid and cost-effective alternatives to experimental testing [1]. However, these computational approaches must overcome significant hurdles related to data quality, interspecies variability, and model interpretability to gain regulatory acceptance and widespread adoption.
Core Methodologies for Addressing Data Discrepancies
Quantitative In Vitro-In Vivo Extrapolation (qIVIVE)
Quantitative in vitro-in vivo extrapolation (qIVIVE) represents a fundamental methodology for bridging the gap between laboratory assays and human physiological responses. This approach is essential for extrapolating an in vitro-based point of departure to a human equivalent dose using kinetic models [66]. When using human cells, qIVIVE can directly extrapolate to human equivalent doses, thereby avoiding interspecies extrapolation challenges that arise with animal cells [66].
The qIVIVE process involves multiple critical steps: (1) mechanistically and quantitatively linking key events to in vivo adverse health effects; (2) selecting appropriate and validated in vitro assays; (3) measuring concentration-response relationships; (4) calculating appropriate concentration metrics; (5) selecting and parametrizing physiologically based kinetic (PBK) models; (6) selecting appropriate model compartments; (7) choosing appropriate dose metrics; (8) performing reverse dosimetry to obtain human external doses related to in vitro concentrations; and (9) selecting appropriate benchmark responses for benchmark dose modeling [66]. Each step introduces potential uncertainties that must be carefully addressed to ensure reliable predictions.
Machine Learning and Artificial Intelligence Approaches
Machine learning (ML) and artificial intelligence (AI) have revolutionized ADMET prediction by enhancing compound optimization, predictive analytics, and molecular modeling [11]. These technologies have demonstrated significant promise in predicting key ADMET endpoints, outperforming some traditional quantitative structure-activity relationship (QSAR) models [65]. ML-based approaches provide rapid, cost-effective, and reproducible alternatives that integrate seamlessly with existing drug discovery pipelines [33].
Core AI algorithms including support vector machines, random forests, graph neural networks, and transformers have been successfully applied to molecular representation, virtual screening, and ADMET property prediction [11]. Deep learning architectures, such as message-passing neural networks and multitask learning frameworks, have shown remarkable capabilities in modeling complex activity landscapes by leveraging large-scale compound databases [33] [14]. These approaches are particularly valuable for addressing data variability through their ability to identify complex patterns in heterogeneous datasets.
Quantum Mechanics/Molecular Mechanics (QM/MM) Methods
Quantum mechanics and molecular mechanics simulations provide fundamental insights into molecular interactions that underlie ADMET properties. Thanks to significant advances in computer speed and new software, quantum mechanics calculations are now used regularly to study drug-related problems, including exploring enzyme-inhibitor interactions, predicting reactivity and stability, and predicting routes of biotransformation [1].
QM/MM approaches have been particularly valuable for understanding metabolic processes mediated by cytochrome P450 (CYP) enzymes, which are responsible for the biotransformation of approximately three-quarters of drugs that undergo metabolism before elimination [1]. These methods have helped resolve controversies about enzyme reactivity and reaction mechanisms, such as the role of heme propionates in P450cam catalysis [1]. By providing atomistic-level insights, QM/MM methods help address fundamental questions about metabolic differences between species.
Comparative Analysis of Computational Approaches
Table 1: Performance Comparison of Key ADMET Prediction Methods
| Methodology | Primary Application | Prediction Accuracy | Computational Cost | Key Limitations |
|---|---|---|---|---|
| qIVIVE with PBK models | Extrapolation from in vitro to in vivo | High variability (11-27% within 2-fold without correction) [67] | Medium to High | Requires extensive parameterization; sensitive to model assumptions |
| Machine Learning (RF, SVM, GNN) | End-to-end ADMET prediction | 45-57% within 2-fold for optimized models [67] [14] | Low to Medium | Dependent on data quality and quantity; black-box concerns |
| QM/MM Simulations | Metabolic pathway prediction | Atomistic accuracy for specific enzymes [1] | Very High | Limited to specific enzymes and small molecule sets |
| Deep Learning (MPNN, Transformers) | Complex structure-property relationships | Outperforms traditional QSAR in many benchmarks [33] [14] | Medium to High | Data hunger; limited interpretability |
Table 2: Empirical Scaling Factors for Improving Hepatic Clearance Predictions
| In Vitro System | Geometric Mean Fold Error (gmfe) | % within 2-fold (Uncorrected) | % within 2-fold (with ESF) |
|---|---|---|---|
| Human Hepatocytes | 10.4 [67] | 27% [67] | 57% [67] |
| Human Liver S9 | 5.0 [67] | 19% [67] | 45% [67] |
| Human Liver Cytosols | 5.6 [67] | 11% [67] | 46% [67] |
Experimental Protocols and Methodologies
qIVIVE Implementation Protocol
The implementation of qIVIVE follows a standardized workflow that begins with the selection of appropriate in vitro assays using human cell lines, such as HepaRG cells for liver steatosis assessment [66]. The protocol involves measuring intracellular concentrations or unbound concentrations in the medium related to the nominal concentrations used in the in vitro assay. Researchers then select and parameterize a physiologically based kinetic model, choosing the most appropriate compartments (e.g., liver or venous blood) for extrapolation.
The critical reverse dosimetry step involves selecting relevant exposure durations and types (e.g., daily bolus doses for prolonged periods) to obtain human external doses related to the in vitro concentrations [66]. Finally, appropriate benchmark responses are selected for benchmark dose modeling to obtain in vitro-based human equivalent doses. This protocol requires careful attention to uncertainty quantification at each step, particularly for extrapolations from short-term in vitro exposure to chronic in vivo exposure and from average human responses to sensitive individuals [66].
Machine Learning Model Development Workflow
The development of machine learning models for ADMET prediction follows a rigorous workflow that begins with comprehensive data cleaning and standardization [14]. This includes removing inorganic salts and organometallic compounds, extracting organic parent compounds from salt forms, adjusting tautomers for consistent functional group representation, canonicalizing SMILES strings, and de-duplicating entries with inconsistent measurements.
Following data preparation, researchers systematically evaluate multiple feature representations including RDKit descriptors, Morgan fingerprints, and deep neural network embeddings [14]. Model selection involves comparing classical algorithms (Support Vector Machines, Random Forests) with more recent neural architectures (Message Passing Neural Networks) using cross-validation with statistical hypothesis testing. The optimized models are then evaluated in practical scenarios where models trained on one data source are tested on datasets from different sources to assess generalizability [14].
Hybrid AI-PBPK Modeling Framework
A emerging methodology combines AI-based feature extraction with traditional PBPK modeling to enhance prediction accuracy while maintaining physiological relevance. This framework uses deep learning architectures such as Mol2Vec to encode molecular substructures into high-dimensional vectors, which are combined with selected chemical descriptors and processed through multilayer perceptrons to predict human-specific ADMET endpoints [46]. The AI component handles complex structure-property relationships while the PBPK model provides physiological constraints and enables extrapolation to human doses.
This hybrid approach supports multi-task learning across multiple ADMET endpoints, allowing the capture of broader interdependencies that simpler systems often miss [46]. The framework includes multiple variants optimized for different virtual screening contexts, ranging from fast models relying solely on substructure embeddings to more accurate but computationally intensive versions incorporating curated molecular descriptors [46].
Visualization of Methodological Workflows
ADMET Prediction Methodology Selection Workflow
Quantitative In Vitro-In Vivo Extrapolation (qIVIVE) Process
The Scientist's Toolkit: Essential Research Reagents and Platforms
Table 3: Key Computational Tools and Platforms for ADMET Prediction
| Tool/Platform | Type | Primary Function | Key Features |
|---|---|---|---|
| Receptor.AI ADMET | Commercial Platform | Multi-endpoint ADMET prediction | Mol2Vec embeddings, 38 human-specific endpoints, multi-task learning [46] |
| Chemprop | Open-source Software | Message-passing neural networks | Molecular property prediction, interpretable latent representations [14] |
| ADMETlab 2.0/3.0 | Web Platform | Integrated ADMET predictions | User-friendly interface, partial multi-task learning [65] [46] |
| Therapeutics Data Commons (TDC) | Data Resource | Curated ADMET benchmarks | Standardized datasets, leaderboard for model comparison [14] |
| RDKit | Cheminformatics Toolkit | Molecular descriptor calculation | Morgan fingerprints, RDKit descriptors, structural standardization [14] |
| Monte Carlo Risk Assessment (MCRA) | Risk Assessment Platform | Probabilistic risk modeling | qIVIVE module, uncertainty quantification [66] |
The landscape of computational ADMET prediction is rapidly evolving, with multiple methodologies now available to address the persistent challenges of interspecies differences and data variability. Our comparative analysis demonstrates that while each approach has distinct strengths and limitations, the integration of multiple strategies often yields the most robust predictions. The emergence of AI-powered platforms that combine multi-task deep learning with traditional physicochemical principles represents a particularly promising direction for the field.
Future advancements will likely focus on enhancing model interpretability, improving data quality through standardized curation practices, and developing more sophisticated uncertainty quantification methods. As regulatory agencies like the FDA formally include AI-based toxicity models under their New Approach Methodologies framework, the validation and standardization of these computational approaches will become increasingly critical [46]. By systematically addressing data discrepancies through the integrated application of qIVIVE, machine learning, and molecular modeling, researchers can significantly improve the accuracy of ADMET predictions and reduce late-stage attrition in drug development.
The emergence of proteolysis-targeting chimeras (PROTACs) represents a paradigm shift in therapeutic modalities, moving beyond traditional inhibition to targeted protein degradation [68]. These heterobifunctional molecules recruit the ubiquitin-proteasome system to degrade disease-causing proteins, offering promise for targeting previously "undruggable" targets [68] [69]. However, their unique molecular architecture introduces substantial Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) challenges that complicate development and require specialized optimization strategies [70] [71].
PROTACs typically violate Lipinski's Rule of Five, with molecular weights ranging from 600-1,000 Da and high polar surface areas that inherently limit oral bioavailability and membrane permeability [68] [71]. Their bifunctional nature—consisting of a target protein ligand, E3 ligase ligand, and connecting linker—creates complex physicochemical properties that often fall outside the applicability domain of traditional ADMET assays [72] [71]. Furthermore, their catalytic mechanism of action and susceptibility to the "hook effect" create non-linear pharmacokinetic-pharmacodynamic (PK/PD) relationships not observed with conventional small molecules [68]. This guide systematically compares ADMET properties and optimization strategies for PROTACs against traditional small molecules, providing experimental frameworks for their characterization within the broader context of validating in silico ADMET predictions.
Comparative ADMET Properties: PROTACs vs. Traditional Small Molecules
Table 1: Comparative ADMET Properties of PROTACs vs. Traditional Small Molecules
| ADMET Parameter | Traditional Small Molecules | PROTACs | Key Implications |
|---|---|---|---|
| Molecular Weight | Typically <500 Da | 600-1,000+ Da [68] [71] | Challenges for oral absorption and permeability |
| Solubility | Predictable with standard assays | Often poor; requires biorelevant media (FaSSIF/FeSSIF) [70] [72] | Impacts formulation strategy and bioavailability |
| Permeability | Assessable via PAMPA | Traditional PAMPA often fails; requires cell-based systems (Caco-2, MDR1-MDCK) [70] | Limits cellular uptake and intracellular activity |
| Protein Binding | Moderate to high; predictable | Very high; difficult to accurately measure [70] [71] | Complicates bioanalysis and pharmacological activity prediction |
| Metabolic Stability | Primarily CYP-mediated | Complex metabolism, especially at linker regions; CYP3A4 common [70] [68] | Affects clearance predictions and potential DDI |
| Clearance Pathways | Hepatic metabolism, renal excretion | Hepatic metabolism predominates; limited biliary/renal clearance [71] | Influences PK predictions across species |
| Cellular Uptake | Passive diffusion often sufficient | Often limited due to size and polarity [68] | Restricts degradation efficiency despite extracellular exposure |
Experimental Protocols for PROTAC ADMET Profiling
Solubility Assessment in Biorelevant Media
Traditional aqueous solubility assays frequently fail to predict the in vivo performance of PROTACs due to their complex physicochemical properties [72]. Instead, biorelevant media simulating intestinal conditions provide more clinically relevant data.
Protocol:
- Preparation of Biorelevant Media: Create FaSSIF (Fasted State Simulated Intestinal Fluid) containing 3 mM sodium taurocholate and 0.75 mM lecithin, and FeSSIF (Fed State Simulated Intestinal Fluid) with higher surfactant content [70].
- Sample Incubation: Add PROTAC candidate to both media at multiple concentrations (e.g., 1-100 µM) and incubate at 37°C with continuous shaking for 2-24 hours.
- Separation: Centrifuge samples at 15,000 × g for 10 minutes to separate undissolved compound.
- Quantification: Analyze supernatant using HPLC-UV or LC-MS/MS to determine dissolved compound concentration [70].
- Data Interpretation: Compare solubility in FaSSIF vs. FeSSIF to anticipate food effects and guide formulation strategy.
Permeability Assessment
Traditional parallel artificial membrane permeability assay (PAMPA) models often fail for PROTACs, necessitating more biologically complex systems [70].
Protocol (Caco-2 Assay):
- Cell Culture: Seed Caco-2 cells at high density (e.g., 100,000 cells/insert) on Transwell inserts and culture for 21-28 days to ensure full differentiation and tight junction formation.
- Dosing: Prepare PROTAC solutions in transport buffer (e.g., HBSS) at multiple concentrations (1-50 µM). Apply to donor compartment (apical for A→B transport, basal for B→A transport).
- Sampling: Collect samples from receiver compartment at regular intervals (e.g., 30, 60, 90, 120 minutes).
- Analysis: Quantify PROTAC concentrations using LC-MS/MS with optimized ionization parameters to address signal complexity [70] [68].
- Efflux Ratio Calculation: Determine Papp (apparent permeability) in both directions and calculate efflux ratio (B→A/A→B). Ratios >2.5 suggest active efflux transport [72].
Plasma Protein Binding Determination
PROTACs frequently exhibit very high plasma protein binding, complicating accurate measurement of unbound fraction (fu) [71]. Traditional methods like rapid equilibrium dialysis (RED) often fail with highly lipophilic PROTACs [71].
Protocol (Ultracentrifugation with Diluted Plasma):
- Sample Preparation: Dilute plasma (1:10) with phosphate buffer to reduce nonspecific binding while maintaining protein integrity [70].
- Incubation: Spike PROTAC into diluted plasma to achieve therapeutic relevant concentration (e.g., 1-5 µM) and incubate at 37°C for 15 minutes.
- Ultracentrifugation: Centrifuge at 436,000 × g for 5 hours at 37°C using an Optima Max-XP ultracentrifuge with TLA-55 rotor [70].
- Analysis: Carefully collect supernatant and analyze using LC-MS/MS with appropriate internal standards.
- Calculation: Determine fu (%) by comparing supernatant concentration to original spiked concentration, with correction for dilution factors.
Metabolic Soft Spot Identification
PROTACs demonstrate complex metabolic pathways, with particular vulnerability at linker regions [68]. Systematic metabolite identification is crucial for guiding structural optimization.
Protocol:
- Incubation Systems Preparation: Use multiple in vitro systems: liver microsomes (for CYP-mediated oxidation), hepatocytes (full enzyme complement), liver S9 fractions (oxidative and conjugative metabolism), and cytosolic fractions (non-CYP enzymes like aldehyde oxidase) [68].
- Incubation: Incubate PROTAC (1-10 µM) with each system at 37°C for 0-120 minutes with NADPH cofactor for oxidative metabolism.
- Sample Processing: Terminate reactions with acetonitrile at multiple time points, centrifuge, and analyze supernatants.
- Metabolite Identification: Use high-resolution LC-MS/MS to identify and semi-quantify metabolites. Focus particularly on linker region modifications [68].
- Data Integration: Prioritize metabolic soft spots based on metabolite abundance and structural importance to guide linker optimization or functional group protection.
Diagram 1: PROTAC ADMET Profiling and Optimization Workflow. This diagram illustrates the interconnected nature of PROTAC structure, ADMET assessment methods, and resulting optimization strategies. Red arrows indicate challenge areas, while green arrows show corresponding solutions.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for PROTAC ADMET Profiling
| Reagent/Assay System | Function in PROTAC ADMET | Key Considerations |
|---|---|---|
| Biorelevant Media (FaSSIF/FeSSIF) | Predicts solubility under physiological conditions | Provides more clinically relevant data than aqueous buffers [70] |
| Caco-2/MDR1-MDCK Cells | Assesses permeability and efflux | More predictive than PAMPA for large, complex molecules [70] |
| Low-Binding Labware | Prevents nonspecific binding during bioanalysis | Critical for accurate quantification; use polypropylene with additives [68] |
| Human Hepatocytes | Evaluates comprehensive metabolic stability | Provides full enzyme complement versus limited microsomal systems [68] |
| Liver S9 Fractions | Identifies oxidative and conjugative metabolism | Broader metabolic coverage than microsomes alone [68] |
| Tween-20/CHAPS | Additives to reduce nonspecific binding | Improve recovery in protein binding and bioanalysis assays [68] |
| Fresh/Frozen Plasma | Plasma protein binding assessment | Evaluate stability in different matrices; fresh may better reflect in vivo conditions [68] |
Advanced PROTAC Optimization Strategies
Pro-PROTAC/Prodrug Approaches
The development of PROTAC prodrugs (pro-PROTACs) represents an innovative strategy to overcome inherent limitations of conventional PROTACs [73]. These temporarily inactivated derivatives address challenges related to precision targeting, duration of action, and tissue-specific delivery [73].
Photocaged PROTACs (opto-PROTACs): These incorporate photolabile groups (e.g., 4,5-dimethoxy-2-nitrobenzyl/DMNB) on critical functional elements—typically the E3 ligase binding moiety (e.g., the glutarimide -NH of CRBN ligands or hydroxyproline of VHL ligands) or occasionally the target protein ligand [73]. Installation of these cages disrupts essential hydrogen bonding interactions required for ternary complex formation, rendering the PROTAC biologically inert until precise spatial and temporal activation with specific wavelength light (commonly 365 nm UV) removes the protecting group [73]. This approach enables unprecedented control over protein degradation, allowing researchers to investigate dynamic cellular processes and potentially reduce off-target effects in therapeutic contexts.
Other Pro-PROTAC Modalities: Beyond light activation, various stimulus-responsive pro-PROTACs are emerging, including enzyme-cleavable systems that leverage disease microenvironment factors (e.g., elevated phosphatase or protease activity) for selective activation [73]. These advanced prodrug strategies aim to enhance the therapeutic index by concentrating active PROTAC delivery specifically to pathological tissues while minimizing exposure to healthy cells [73].
Formulation Technologies for Enhanced Delivery
Advanced formulation strategies play a crucial role in overcoming the inherent physicochemical challenges of PROTACs [68].
Lipid-Based Delivery Systems: These include self-emulsifying drug delivery systems (SEDDS) and lipid nanoparticles that enhance solubility and lymphatic absorption, bypassing first-pass metabolism [68] [71].
Amorphous Solid Dispersions (ASDs): Spray-dried or hot-melt extruded dispersions can significantly increase dissolution rates and maintain supersaturation through polymer-based stabilization [68].
Nanoparticle-PROTAC Conjugates: Inorganic or polymeric nanoparticles can be functionalized with PROTACs to improve tissue targeting and cellular uptake, particularly valuable for otherwise impermeable targets [69].
Machine Learning in PROTAC Optimization
Artificial intelligence and machine learning are increasingly applied to accelerate PROTAC development and optimize ADMET properties [33]. These computational approaches include:
Predictive Linker Design: Models like AIMLinker (deep encoder-decoder neural network) and ShapeLinker (reinforcement learning-based) generate novel, synthetically accessible linker structures optimized for degradation efficiency and physicochemical properties [73].
ADMET Property Prediction: Graph neural networks (GNNs) and ensemble learning methods analyze complex structure-property relationships to forecast permeability, metabolic stability, and toxicity endpoints, enabling virtual screening of PROTAC candidates before synthesis [33].
Ternary Complex Modeling: Platforms such as DeepPROTAC process ligand and binding pocket information through graph convolutional networks (GCNs) to predict degradation efficacy and inform rational design [73].
PROTACs present a distinct set of ADMET challenges that necessitate specialized assay modifications and optimization strategies diverging from traditional small molecule approaches. Their complex molecular architecture demands biorelevant solubility assessment, cell-based permeability models, and comprehensive metabolic soft spot identification. The promising clinical progression of candidates like vepdegestrant (ARV-471) demonstrates that despite physicochemical properties beyond Rule-of-five, viable oral PROTAC drugs are achievable through strategic optimization of linkers, utilization of advanced formulations, and implementation of prodrug approaches [68] [73]. As the field advances, the integration of machine learning with multi-omics validation and exploration of novel E3 ligases will further accelerate the development of this transformative therapeutic modality, ultimately expanding the druggable proteome for patients with limited treatment options.
Improving Oral Bioavailability Predictions Using Gut-Liver Model Systems
Oral bioavailability, defined as the fraction of an orally administered drug that reaches systemic circulation, is a critical pharmacokinetic (PK) parameter optimized by drug developers to establish safe and effective therapeutic dosages [74] [75]. Its optimization is paramount for drug efficacy, as insufficient oral bioavailability can stall the development of oral therapeutics [74]. Bioavailability (F) is determined by the interplay of three key components: the fraction absorbed (Fa), the fraction escaping gut wall elimination (Fg), and the fraction escaping hepatic elimination (Fh), summarized by the equation F = Fa × Fg × Fh [74] [75].
The drug discovery process has traditionally lacked robust in vitro assays that can simultaneously profile the contributions of the gut and liver to overall bioavailability [74] [75]. Conventional approaches use isolated systems—such as Caco-2 cells for gut absorption and liver microsomes or suspension hepatocytes for hepatic metabolism—but these operate in isolation and fail to capture the complex, integrated physiology of first-pass metabolism [74] [76]. Furthermore, traditional in vivo animal models are poor quantitative predictors of human bioavailability, with one seminal study of 184 compounds showing no absolute correlation between human and any individual species (R² ≈ 0.34) [74] [76]. This translational gap underscores the need for more human-relevant approaches. Microphysiological systems (MPS), also known as organ-on-a-chip platforms, have emerged as advanced in vitro tools that fluidically link multiple organ tissues to better mimic human physiology [74]. This guide objectively compares the performance of a next-generation primary human Gut-Liver MPS against established alternatives, providing experimental data to frame these advances within the broader thesis of validating in silico ADMET predictions with high-quality in vitro data.
Comparative Analysis of Model Systems for Bioavailability Prediction
The following section provides a detailed, data-driven comparison of the available model systems, highlighting the performance and limitations of each approach.
Table 1: Comparison of Model Systems for Oral Bioavailability Prediction
| Model System | Key Features | Advantages | Limitations / Performance Data |
|---|---|---|---|
| Primary Human Gut-Liver MPS [74] [75] [76] | - Fluidically linked primary human jejunum epithelium and primary human hepatocytes (PHHs)- Dual-organ supporting media- Mimics both oral and intravenous dosing routes | - Fully human, primary cell-based- Maintains metabolic functionality of both tissues- Enables mechanistic modeling of Fa, Fg, Fh- Provides human-relevant data for PBPK modeling | Overcomes key Caco-2 limitations: Demonstrates superior predictive capacity for drugs subject to intestinal metabolism (e.g., CYP3A4 substrates like midazolam) [75] [76]. |
| Caco-2/Liver MPS [74] [76] | - Fluidically linked Caco-2 intestinal model and PHHs- Chemically defined co-culture media | - More integrated than isolated assays- Improved physiological relevance over single-organ systems | Limited by Caco-2 biology: Absent or low levels of key drug-metabolizing enzymes (e.g., CYP3A4) and transporters hinder accurate Fg prediction [74] [76]. |
| Traditional In Vitro Assays (Isolated Caco-2, liver microsomes) [74] [76] | - Simple, well-established protocols- Used in isolation | - Low-cost, high-throughput- Standardized for early-stage screening | Lack integration: Assess gut absorption and liver metabolism in isolation, failing to capture the interplay of first-pass metabolism [74]. Poor predictors of human Fg and Fh [74]. |
| QSAR/Machine Learning Models [77] [78] | - In silico prediction using molecular structure and descriptors- Algorithms include Random Forest, CatBoost, and graph-based deep learning | - Extremely rapid and cheap- Useful for virtual screening of large compound libraries | Limited predictive performance: Best regression models for bioavailability (F%) show modest performance (e.g., Q²F₃ of 0.34 for a large dataset of 1,712 chemicals) [78]. Performance is hampered by data quality and the "black box" problem [79]. |
| Animal Models [74] [76] | - Provides data within a whole, living organism- Historical "gold standard" for regulatory submissions | - Captures complex systemic physiology | Poor human translatability: Weak quantitative correlation with human bioavailability (R² ~0.34 for 184 compounds) [74] [76]. Ethical concerns and high costs [76]. |
Experimental Data and Workflow from a Primary Human Gut-Liver MPS
Detailed Experimental Protocol
The established protocol for the primary human Gut-Liver MPS provides a framework for generating high-quality, human-relevant data [74].
- Liver MPS Setup: Primary human hepatocytes (PHHs) are seeded at a density of 0.6 × 10⁶ viable cells per well on a collagen-coated scaffold in a multi-chip plate. The system is cultured using a pneumatically driven microfluidic pump that recirculates proprietary Liver Maintenance Medium at a flow rate of 1.0 µL/s. PHH functionality is assessed by quantifying albumin and urea production, lactate dehydrogenase (LDH) release for cell health, and CYP3A4 enzyme activity using a P450-Glo assay [74].
- Primary Gut Model Setup: Human jejunum stem/progenitor cells (RepliGut) are expanded on a Transwell membrane coated with a proprietary hydrogel. Cells are first cultured in RepliGut Growth Medium until confluence, then switched to RepliGut Maturation Medium to promote differentiation and polarization into a functional intestinal epithelial monolayer. Gut barrier integrity is monitored daily via transepithelial electrical resistance (TEER) measurements [74].
- Gut-Liver Coculture Establishment: After the liver microtissues have matured (4 days post-seeding) and the primary gut model has fully differentiated (day 13), the two systems are integrated into a dual-organ plate. The coculture is maintained in two specially formulated media: Gut/Liver Apical Medium (GAM) is added to the apical side of the gut compartment, and Gut/Liver Circulation Medium (GCM) is added to the basolateral side, which fluidically connects the gut and liver compartments under continuous flow [74].
- Dosing and Bioavailability Estimation: The model allows simulation of both oral (via the gut compartment) and intravenous (via the circulation medium) dosing routes. As a case study, the CYP3A4 substrate midazolam was used. Concentrations of the parent drug and its metabolite (1'-hydroxymidazolam) are measured in the system over time. The resulting concentration-time data are integrated with a mechanistic mathematical model to generate organ-specific PK parameters and estimate human oral bioavailability (F) and its components (Fa, Fg, Fh) [74] [75].
Signaling Pathways and Experimental Workflow
The diagram below illustrates the key biological process of first-pass metabolism and the experimental workflow of the Gut-Liver MPS.
The Scientist's Toolkit: Essential Research Reagent Solutions
The successful implementation of advanced MPS models relies on a suite of specialized reagents and tools. The following table details key solutions used in the featured primary Gut-Liver MPS study.
Table 2: Key Research Reagent Solutions for Gut-Liver MPS
| Research Reagent / Tool | Function in the Experiment | Source / Example |
|---|---|---|
| Primary Human Hepatocytes (PHHs) | Forms the 3D liver microtissue responsible for drug metabolism; provides crucial CYP450 enzyme activity (e.g., CYP3A4). | Commercially available from providers like LifeNet Health [74]. |
| RepliGut Planar Jejunum Model | Provides a primary human intestinal epithelial monolayer derived from jejunum stem/progenitor cells; enables accurate modeling of absorption and gut metabolism. | Altis Biosystems (Catalog: RGP-JEJ-PMX) [74]. |
| PhysioMimix Multi-organ System | The core hardware and consumables (e.g., Dual-organ Plate) that provide fluidic flow and environmental control to maintain tissue viability and enable interconnection. | CN Bio Innovations [74] [75]. |
| Gut/Liver Co-culture Media | Specially formulated, proprietary media that maintain the differentiation and metabolic function of both gut and liver tissues in a shared fluidic environment. | CN Bio Innovations (Gut/Liver Apical & Circulation Media) [74]. |
| Mechanistic Mathematical Model | In silico framework that translates concentration-time data from the MPS into organ-specific PK parameters and estimates of human Fa, Fg, Fh, and F. | Developed in conjunction with the experimental MPS [74] [75]. |
The integration of advanced in vitro models like the primary human Gut-Liver MPS with in silico modeling represents a paradigm shift in predicting oral bioavailability. This approach directly addresses the critical translational gap left by traditional animal models and isolated in vitro assays. By providing high-quality, human-relevant data on the key components of bioavailability (Fa, Fg, Fh), these systems offer a powerful means to de-risk drug candidates earlier in the development process and provide more reliable data for refining PBPK models. This progress is a significant step forward in the broader thesis of validating and enhancing in silico ADMET predictions with robust, physiologically relevant in vitro data, ultimately promising to improve the efficiency and success rate of clinical drug development.
Tackling Limitations of Traditional In Vitro Systems with Organ-on-a-Chip Technology
The drug development pipeline has long been hampered by the inadequate predictive power of traditional in vitro systems. Conventional two-dimensional (2D) cell cultures, while useful for basic research, fail to replicate the complex microenvironment of human tissues, leading to poor translatability of results to clinical outcomes [80]. This technological gap contributes significantly to the high attrition rates in drug development, with approximately 30% of drug failures attributed to unforeseen toxicity issues that were not predicted during pre-clinical testing [81]. Organ-on-a-Chip (OOC) technology represents a transformative approach that bridges this critical gap by leveraging microfluidic engineering to create physiologically relevant models that recapitulate the structure and function of human organs.
OOC devices are microfluidic cell culture systems that simulate the activities, mechanics, and physiological responses of entire organs and organ systems. These innovative platforms provide the best of both in vivo and in vitro research by offering human-relevant data while maintaining controlled experimental parameters [80]. The fundamental advantage of OOC technology lies in its ability to utilize microfluidics to ensure a stable concentration gradient, dynamic mechanical stress modeling, and accurate reconstruction of a cellular microenvironment—features entirely absent in traditional static cultures [80]. This capability is particularly valuable for ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) predictions, where accurate simulation of human physiological responses is crucial for validating in silico models and improving drug safety profiling.
Comparative Analysis: OOC vs. Traditional In Vitro Systems
Fundamental Limitations of Traditional In Vitro Models
Traditional 2D cell culture systems suffer from several critical limitations that restrict their predictive capacity. They lack the three-dimensional architecture characteristic of native tissues, which disrupts normal cell-cell and cell-matrix interactions essential for proper cellular differentiation and function [82]. Without fluid flow and mechanical stimulation, these static systems cannot replicate the biomechanical forces that cells experience in vivo, such as shear stress from blood flow or cyclic strain from breathing movements [80] [82]. Furthermore, traditional models fail to establish physiological nutrient and oxygen gradients that drive tissue organization and function, ultimately resulting in poor representation of human physiology and compromised predictability for drug responses [82].
Technical Advantages of Organ-on-a-Chip Systems
Organ-on-a-Chip technology addresses these limitations through sophisticated engineering approaches that better mimic human physiology. The core advantages include:
- Reconstruction of tissue-tissue interfaces: OOC devices can incorporate multiple cell types separated by porous membranes, recreating critical biological barriers like the alveolar-capillary interface in the lung or the intestinal epithelium [80] [82].
- Application of dynamic mechanical forces: These systems can apply physiological relevant mechanical cues, including cyclic strain to simulate breathing in lung chips and fluid flow to mimic blood circulation [80] [83].
- Precise control of microenvironment: Microfluidics enables the establishment of stable concentration gradients and spatiotemporal control over signaling molecules, more accurately representing in vivo conditions [80] [82].
- Integration of multiple organ systems: Multi-organ chips allow for the study of inter-tissue interactions and systemic drug effects, providing insights into metabolite-mediated organ crosstalk [80] [84].
Table 1: Performance Comparison of Traditional In Vitro Models vs. Organ-on-a-Chip Technology
| Parameter | Traditional 2D Models | Organ-on-a-Chip Systems |
|---|---|---|
| Architectural Complexity | Simple monolayer without 3D structure | 3D tissue-like structures with physiological organization |
| Mechanical Forces | Static conditions without flow or strain | Dynamic fluid flow and application of physiological strain |
| Biochemical Gradients | Limited, unstable gradients due to static culture | Precise, stable concentration gradients via microfluidics |
| Tissue-Tissue Interfaces | Difficult to establish and maintain | Reproducible incorporation of tissue barriers |
| Cell-Cell Interactions | Limited to single cell type or simple co-cultures | Complex multi-cellular environments with physiological spatial arrangement |
| Functional Duration | Typically days | Weeks to months of stable culture [85] |
| Predictive Capacity for Human Response | Limited, poor clinical translatability | Improved accuracy for drug absorption, metabolism, and toxicity [84] |
| Systemic Response Modeling | Not possible | Multi-organ interactions possible through linked systems |
Quantitative Performance Comparison in ADMET Applications
The enhanced physiological relevance of OOC systems translates directly to improved performance in key ADMET applications. Recent studies have demonstrated the superior predictive capability of these systems, particularly for drug absorption and metabolism studies.
Table 2: Experimental Performance Data for ADMET Prediction Using Gut/Liver-on-a-Chip Model
| Metric | Traditional In Vitro Methods | Gut/Liver-on-a-Chip | Clinical Data |
|---|---|---|---|
| Midazolam Bioavailability Prediction | Not accurately predictable with isolated systems | 44% (within clinical range) [84] | 30-60% [84] |
| Hepatic Clearance (CLint,liver) | Requires multiple separate assays | Quantified from single experiment [84] | N/A |
| Intestinal Permeability (Papp) | Measured in isolation without first-pass metabolism | Determined with simultaneous gut-liver interaction [84] | N/A |
| Fraction Absorbed (Fa) | Caco-2 models without physiological flow | Calculated with physiological flow conditions [84] | N/A |
| Cost per Compound Screening | $100,000 - $500,000+ (including animal studies) | Significant reduction by reducing animal study reliance [81] [84] | N/A |
| Time for Bioavailability Assessment | Weeks to months (including animal studies) | Days to weeks with integrated system [84] | N/A |
Experimental Protocols and Methodologies
Integrated Gut/Liver-on-a-Chip for Bioavailability Assessment
The PhysioMimix Gut/Liver model developed by CN Bio represents a state-of-the-art experimental platform for predicting human drug bioavailability. This protocol outlines the key methodology for assessing drug absorption and metabolism using this system [84]:
Device Setup and Cell Culture:
- Utilize a microfluidic device with separate but interconnected gut and liver compartments.
- Seed the intestinal compartment with human primary intestinal epithelial cells (or appropriate cell lines) on a porous membrane coated with extracellular matrix to form a differentiated epithelial layer.
- Seed the liver compartment with primary human hepatocytes in a 3D configuration to maintain metabolic function.
- Maintain both tissues under continuous, low-rate perfusion (0.1-10 μL/min) to mimic physiological flow conditions.
- Culture the system for 7-14 days to allow for full tissue maturation and stabilization of metabolic functions before experimentation.
Experimental Dosing and Sampling:
- Introduce the test compound dissolved in fasted or fed state simulated intestinal fluid to the gut compartment inlet.
- Collect effluent from both the gut and liver outlets at predetermined time points (e.g., 0, 1, 2, 4, 8, 12, 24, 48, and 72 hours).
- Analyze samples using LC-MS/MS to quantify parent compound and major metabolites.
- Monitor tissue viability and integrity throughout the experiment using transepithelial electrical resistance (TEER) measurements for the gut compartment and albumin/urea production for the liver compartment.
Data Analysis and Computational Integration:
- Measure compound concentration over time in both compartments to establish absorption and metabolism profiles.
- Apply mathematical modeling to quantify key pharmacokinetic parameters including intrinsic hepatic clearance (CLint,liver), gut clearance (CLint,gut), apparent permeability (Papp), and efflux ratio (Er).
- Use Bayesian methods to determine confidence intervals for each parameter.
- Calculate the components of bioavailability: Fa (fraction absorbed), Fg (fraction escaping gut metabolism), and Fh (fraction escaping hepatic metabolism).
- Determine overall oral bioavailability (F) as the product of these three components: F = Fa × Fg × Fh.
Bone Marrow-on-a-Chip for Toxicity Assessment
The Bone Marrow-on-a-Chip platform provides a human-relevant model for predicting myelosuppression, a common side effect of chemotherapeutic agents and other drugs. The experimental protocol includes [83]:
Device Fabrication and Cellularization:
- Fabricate a microfluidic device with two parallel channels separated by a porous membrane.
- Seed the vascular channel with human endothelial cells to recreate a blood vessel-like structure.
- Fill the parallel channel with a fibrin gel matrix seeded with CD34⁺ hematopoietic stem and progenitor cells along with stromal cells to recreate the bone marrow niche.
- Apply continuous perfusion at physiologically relevant flow rates to deliver nutrients and mimic circulation.
- Culture the system for 3-4 weeks to allow for differentiation and maturation of multiple blood cell lineages (myeloid, erythroid, megakaryocytic).
Toxicity Testing:
- Expose the mature bone marrow chip to clinically relevant concentrations of the test compound(s) for single or repeated doses.
- Monitor lineage-specific depletion through regular sampling of the effluent and analysis of cellular composition.
- Assess viability and functional capacity of hematopoietic cells through colony-forming unit assays.
- For patient-specific toxicity modeling, utilize CD34⁺ cells isolated from individuals with specific conditions (e.g., Shwachman-Diamond syndrome) to assess compound effects on vulnerable populations.
Integrated In Silico and OOC Workflow for ADMET Prediction
The combination of OOC experimental data with computational modeling represents a powerful approach for enhancing ADMET predictions [84]:
Experimental Phase:
- Conduct OOC experiments as described in section 3.1 to generate time-concentration data for parent drug and metabolites.
- Determine key parameters such as metabolic rates, permeability, and tissue partitioning coefficients.
Computational Modeling Phase:
- Develop mathematical models describing drug movement and metabolism throughout the OOC system.
- Generate multiple feasible models with distinct assumptions about rate-limiting steps and mechanisms.
- Fit all candidate models to the experimental dataset using appropriate statistical methods.
- Select the best-performing model based on pre-defined performance criteria (e.g., Akaike Information Criterion, Bayesian Information Criterion).
- Extract confidence intervals for key ADME parameters using Bayesian methods.
Prediction and Validation:
- Use the optimized model parameters to predict human pharmacokinetic parameters and oral bioavailability.
- Compare predictions with existing clinical data to validate the approach.
- Utilize the parameters to inform physiologically based pharmacokinetic (PBPK) models for first-in-human dose predictions.
Key Signaling Pathways in Organ-on-a-Chip Systems
OOC technology enables the recapitulation of critical signaling pathways that maintain tissue function and mediate drug responses. Understanding these pathways is essential for proper model design and interpretation of experimental results.
The diagram above illustrates key signaling pathways that are activated and maintained in OOC systems, enabling physiological function and responses:
TGF-β/SMAD Pathway: This pathway regulates cell growth, differentiation, and apoptosis in various tissues. In multi-organ systems, TGF-β1 has been shown to coordinate responses across different tissue types [86]. The pathway is particularly important in maintaining epithelial barrier function and mediating tissue-specific responses to injury or toxic insults.
EGF Receptor Pathway: Epidermal growth factor receptor signaling plays a crucial role in cell proliferation, survival, and differentiation. In lung-on-a-chip models, mechanical strain has been shown to modulate EGFR signaling, which subsequently affects cellular responses to therapeutics, including tyrosine kinase inhibitors in cancer models [86].
HIF-1α Oxygen Sensing Pathway: The establishment of physiological oxygen gradients in OOC systems activates hypoxia-inducible factors, which regulate metabolism, angiogenesis, and cell fate decisions. This pathway is essential for proper zonation in liver models and metabolic function.
CXCR4/CXCL12 Homing Axis: In bone marrow-on-a-chip models, this chemokine signaling pathway maintains stem cell niches and regulates hematopoietic differentiation, enabling long-term culture of functional bone marrow tissue [83].
These pathways, often disrupted in traditional 2D cultures, remain functional in OOC platforms due to the presence of physiological mechanical forces, proper cell-cell interactions, and biomimetic microenvironmental cues.
Essential Research Reagent Solutions for OOC Technology
Successful implementation of OOC technology requires specialized reagents and materials that support the complex culture environments. The table below details key solutions used in advanced OOC systems.
Table 3: Essential Research Reagent Solutions for Organ-on-a-Chip Applications
| Reagent/Material | Function | Application Examples | Key Considerations |
|---|---|---|---|
| Polydimethylsiloxane (PDMS) | Primary material for chip fabrication; transparent, gas-permeable, flexible | Universal application for most OOC devices | Potential for small molecule absorption; surface treatment often required [80] |
| Extracellular Matrix (ECM) Hydrogels | Provide 3D scaffolding for cells; mimic native tissue microenvironment | Basement membrane mimics (Matrigel), collagen, fibrin matrices | Tissue-specific formulations enhance physiological relevance [80] |
| Primary Human Cells | Provide human-relevant responses; maintain donor-specific phenotypes | Hepatocytes, intestinal epithelial cells, endothelial cells | Limited availability; donor-to-donor variability [84] |
| Induced Pluripotent Stem Cells (iPSCs) | Patient-specific cells that can differentiate into multiple cell types | Disease modeling, personalized medicine applications | Differentiation efficiency and maturation time vary [83] |
| Pluronic Acid | Surface passivation to prevent unwanted cell attachment | 3D spheroid and organoid cultures | Critical for maintaining 3D architecture in suspension cultures [80] |
| Tissue-Specific Media Formulations | Support viability and function of specialized cell types | Defined media for liver, gut, neural, etc. cultures | Often require optimization for microfluidic environments [85] |
| Oxygen-Sensitive Sensors | Monitor oxygen tension in microenvironments | Real-time metabolic assessment | Enables validation of physiological gradient formation [82] |
Organ-on-a-Chip technology represents a significant advancement over traditional in vitro systems, offering unprecedented physiological relevance for ADMET prediction and drug development. By incorporating dynamic fluid flow, physiological mechanical forces, and complex tissue-tissue interfaces, OOC platforms bridge the critical gap between conventional cell culture and human physiology. The integration of OOC-derived data with computational modeling creates a powerful framework for validating in silico ADMET predictions, ultimately reducing the reliance on animal models and improving the efficiency of drug development [84].
The case studies and experimental data presented demonstrate that OOC technology provides more accurate predictions of human pharmacokinetics and toxicity compared to traditional methods. As regulatory agencies like the FDA increasingly accept alternative approaches under initiatives such as the FDA Modernization Act 2.0, the adoption of OOC systems is poised to accelerate [81] [85]. For researchers focused on validating in silico ADMET predictions, OOC technology offers a human-relevant experimental platform for parameter estimation and model validation, potentially transforming the drug development landscape by providing more reliable, clinically translatable data at earlier stages of the pipeline.
Rigorous Validation Frameworks and Comparative Model Analysis
Accurate prediction of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties remains a fundamental challenge in drug discovery, with approximately 40–45% of clinical attrition still attributed to ADMET liabilities [9]. As model performance increasingly becomes limited by data availability and quality rather than algorithms alone, establishing robust validation protocols has emerged as a critical necessity for building trust in predictive models. The field is transitioning from simple hold-out validation toward more sophisticated frameworks that combine rigorous statistical testing with practical applicability assessments [87] [14]. This evolution addresses a recognized shortcoming in many existing approaches where model selection and compound representation choices often lack systematic justification [14]. Within this context, this guide objectively compares current validation methodologies, providing researchers with a structured framework for evaluating ADMET prediction models in both academic and industrial settings.
Comparative Analysis of Validation Frameworks
Multi-Tiered Validation Approach
A comprehensive validation strategy for ADMET prediction models should incorporate multiple assessment tiers, progressing from internal validation to external applicability testing. The most effective frameworks integrate cross-validation with statistical hypothesis testing, add a layer of reliability to model assessments, and finally evaluate performance on truly external datasets from different sources [87] [14]. This progression ensures that models demonstrate not only statistical significance but also practical utility in real-world drug discovery scenarios.
Table 1: Comparison of Validation Approaches for ADMET Predictive Models
| Validation Method | Key Characteristics | Performance Metrics | Advantages | Limitations |
|---|---|---|---|---|
| K-Fold Cross-Validation with Statistical Testing | Multiple data splits with statistical significance assessment of performance differences [87] [14] | Pearson’s r, MAE, RMSE distributions with p-values [87] | More robust model comparison than single hold-out test; accounts for variability across splits [87] | Does not assess performance on novel chemical scaffolds or different experimental conditions |
| Scaffold-Based Splitting | Splits ensure compounds with similar molecular scaffolds are separated between training and test sets [9] [14] | Performance degradation compared to random splits indicates scaffold bias [9] | Tests generalization to novel chemotypes; more realistic for drug discovery [9] | Typically shows lower performance metrics than random splits |
| External Dataset Validation | Evaluation on data from different sources (e.g., public model vs. pharmaceutical company internal data) [87] [19] | Correlation coefficients, MAE on external set [19] | Assesses real-world applicability and transferability [19] | Potential protocol differences between data sources; can be computationally expensive |
| Temporal Validation | Training on older compounds, testing on newer ones [14] | Time-dependent performance degradation | Simulates real-world deployment where future compounds differ from past | Requires timestamped data which may not be available |
Quantitative Performance Benchmarks
Recent benchmarking initiatives provide critical reference points for expected performance across different ADMET endpoints. The Polaris ADMET Challenge revealed that multi-task architectures trained on broader and better-curated data consistently outperformed single-task or non-ADMET pre-trained models, achieving up to 40–60% reductions in prediction error across endpoints including human and mouse liver microsomal clearance, solubility (KSOL), and permeability (MDR1-MDCKII) [9]. These results highlight that data diversity and representativeness, rather than model architecture alone, are often the dominant factors driving predictive accuracy and generalization.
Table 2: Exemplary Model Performance Across ADMET Endpoints Using Rigorous Validation
| ADMET Endpoint | Best-Performing Algorithm | Representation | Performance (Test Set) | External Validation Outcome |
|---|---|---|---|---|
| Caco-2 Permeability | XGBoost [19] | Morgan fingerprints + RDKit2D descriptors [19] | Superior to RF, GBM, SVM, DMPNN, CombinedNet [19] | Retained predictive efficacy on pharmaceutical company internal data [19] |
| Multiple ADMET Properties | DeepDelta (pairwise approach) [88] | D-MPNN architecture processing molecular pairs [88] | Outperformed ChemProp and Random Forest on 70% of benchmarks (Pearson's r) [88] | Superior performance on all external test sets for both Pearson's r and MAE [88] |
| ADMET Classification Tasks | Random Forest [14] | Combination of selected features [14] | Dataset-dependent optimal performance [14] | Performance varies when applied to different data sources [87] |
| HTTK Parameters | Ensemble QSPR Models [89] | Structural descriptors + OPERA physico-chemical predictions [89] | AUC predictions with RMSLE 0.6–0.8 using QSPR values [89] | Performance similar to using in vitro values for PBTK modeling (RMSLE ∼1) [89] |
Experimental Protocols for Robust Validation
Protocol 1: Cross-Validation with Statistical Hypothesis Testing
Methodology: Implement 5 × 10-fold cross-validation (five repeats of ten-fold validation) to generate performance distributions rather than single point estimates [88] [14]. For each cross-validation split, the training data is first divided into train and test sets before creating molecular pairings to prevent data leakage [88]. Apply appropriate statistical tests (e.g., paired t-tests) to the resulting performance distributions to separate real gains from random noise [9] [14].
Key Steps:
- Perform scaffold-based splitting of datasets to ensure structurally dissimilar compounds between training and test sets [9] [14]
- Generate multiple data splits using different random seeds (recommended: 10 splits) [19]
- Train models on each training split and evaluate on corresponding test splits
- Compute performance metric distributions across all splits
- Apply statistical tests to compare model performances
- Benchmark against various null models and noise ceilings [9]
Implementation Considerations: This approach is particularly valuable for identifying statistically significant differences between models or feature representations that might appear similar based on single train-test splits [87]. The protocol should include dataset-specific hyperparameter tuning before evaluation to ensure fair comparisons [14].
Protocol 2: External Validation with Pharmaceutical Industry Data
Methodology: Assess model transferability by evaluating performance on pharmaceutical company internal datasets after training on public data [19]. This validation step is crucial for determining real-world applicability, as public and internal compounds often differ substantially in chemical space and assay protocols.
Key Steps:
- Train models on curated public datasets (e.g., TDC, ChEMBL) [19] [14]
- Apply trained models directly to pharmaceutical company internal datasets without retraining
- Evaluate performance degradation compared to internal cross-validation results
- Analyze chemical space differences between public and internal compounds
- Perform applicability domain analysis to identify compounds outside model coverage [19]
Implementation Considerations: Studies evaluating Caco-2 permeability models found that boosting models like XGBoost retained a degree of predictive efficacy when applied to pharmaceutical industry data, though performance typically decreases compared to internal validation [19]. This protocol is essential for models intended for deployment in drug discovery pipelines.
Figure 1: Comprehensive Workflow for Robust ADMET Model Validation
Essential Research Reagent Solutions
Table 3: Key Research Tools and Resources for ADMET Validation Studies
| Resource Category | Specific Tools & Databases | Primary Function | Application in Validation |
|---|---|---|---|
| Cheminformatics Toolkits | RDKit [19] [14], descriptastorus [19] | Molecular standardization, fingerprint generation, descriptor calculation | Data preprocessing, feature generation, molecular representations |
| Benchmark Datasets | Therapeutics Data Commons (TDC) [87] [14] [22], PharmaBench [22], ChEMBL [88] [22] | Curated ADMET property measurements | Training and baseline evaluation; external test sets |
| Machine Learning Frameworks | Scikit-learn [88] [14], ChemProp [88] [14], XGBoost [19] [14] | Model implementation and training | Building predictive models with various algorithms |
| Statistical Analysis Tools | SciPy, custom hypothesis testing implementations [87] [14] | Statistical significance testing | Comparing model performance distributions |
| Commercial Platforms | ADMET Predictor [90], Opera [89] | Proprietary prediction algorithms | Benchmarking against established tools |
The establishment of robust validation protocols represents a critical advancement in the field of in silico ADMET prediction. Through the systematic implementation of cross-validation with statistical testing, scaffold-based splits, and external validation, researchers can now develop models with demonstrated generalizability rather than just optimal performance on specific benchmarks. The comparison of methodologies reveals that while no single approach guarantees success, frameworks that incorporate multiple validation tiers consistently provide more reliable assessments of real-world utility. As federated learning and other collaborative approaches continue to expand the chemical space accessible for model training [9], these rigorous validation protocols will become increasingly essential for translating technical advances into practical improvements in drug discovery efficiency. Future methodology development should focus on standardizing these validation approaches across the research community to enable more meaningful comparisons between studies and accelerate the adoption of high-performing models in pharmaceutical R&D.
The optimization of Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties plays a pivotal role in drug discovery, directly influencing a drug's efficacy, safety, and ultimate clinical success [22]. In silico ADMET prediction platforms have emerged as crucial tools for providing early assessment of these properties, helping researchers prioritize compounds with favorable pharmacokinetic profiles and minimize late-stage failures [22] [91]. The landscape of these tools is broadly divided into two categories: open-source platforms, often developed through academic initiatives, and commercial suites backed by specialized software companies. This guide provides a comparative analysis of both approaches, focusing on their performance validation against experimental data to assist researchers, scientists, and drug development professionals in selecting appropriate tools for their specific contexts. The validation of these computational predictions against in vitro data remains a critical step in building confidence for their application in decision-making processes [91].
Key ADMET Properties and Predictive Modeling Approaches
Core ADMET Properties for Prediction
Computational ADMET prediction encompasses a wide range of physicochemical (PC) and toxicokinetic (TK) properties. The table below summarizes the most common endpoints and their implications in drug discovery.
Table 1: Key ADMET Properties and Their Significance in Drug Discovery
| Property Category | Specific Property | Abbreviation | Unit/Format | Role in Drug Discovery |
|---|---|---|---|---|
| Physicochemical (PC) | Octanol/Water Partition Coefficient | LogP | Adimensional | Predicts lipophilicity; affects membrane permeability |
| Water Solubility | LogS | log mol/L | Impacts dissolution and bioavailability | |
| Acid Dissociation Constant | pKa | Adimensional | Influences ionization state, solubility, and permeability | |
| Melting Point | MP | °C | Affects compound stability and crystallinity | |
| Toxicokinetic (TK) | Caco-2 Permeability | Caco-2 | log cm/s | Models human intestinal absorption |
| Blood-Brain Barrier Permeability | BBB | Categorical (Yes/No) | Predicts central nervous system exposure | |
| Fraction Unbound (Plasma) | FUB | Fraction (%) | Indicates amount of drug available for activity | |
| P-glycoprotein Substrate | Pgp.sub | Categorical (Yes/No) | Identifies compounds affected by efflux transporters | |
| Human Intestinal Absorption | HIA | Categorical (e.g., HIA >30%) | Predicts oral bioavailability potential |
Foundational Modeling Approaches
Most ADMET prediction tools, whether open-source or commercial, are built upon Quantitative Structure-Activity Relationship (QSAR) models [91]. These models correlate molecular descriptors derived from a compound's structure with its experimental biological activity or property. The predictive performance is heavily dependent on the quality, size, and chemical diversity of the training data. More recently, Artificial Intelligence (AI) and Machine Learning (ML) have been increasingly integrated into these platforms, enhancing their predictive accuracy and enabling the handling of more complex, non-linear relationships [92] [93]. Commercial tools often incorporate proprietary AI/ML algorithms trained on large, curated datasets, some of which have ranked #1 in independent peer-reviewed comparisons [94].
Comparative Performance Analysis
Quantitative Performance Benchmarking
Independent benchmarking studies are essential for objectively evaluating predictive performance. One comprehensive review evaluated twelve software tools (a mix of open-source and commercial) for predicting 17 PC and TK properties using 41 curated external validation datasets [91]. The results provide a direct comparison of predictive accuracy.
Table 2: External Validation Performance of Computational ADMET Tools [91]
| Property Type | Metric | Average Performance | Performance Insight |
|---|---|---|---|
| Physicochemical (PC) Properties | R² (Regression) | 0.717 (Average) | Models for PC properties generally showed strong predictive performance. |
| Toxicokinetic (TK) Properties | R² (Regression) | 0.639 (Average) | TK regression models showed good but slightly lower performance than PC models. |
| Toxicokinetic (TK) Properties | Balanced Accuracy (Classification) | 0.780 (Average) | Classification models for endpoints like BBB permeability and HIA were robust. |
The study concluded that the majority of the selected tools demonstrated adequate predictive performance, with several emerging as recurring optimal choices across different properties [91]. It emphasized that the best-performing models could be proposed as robust computational tools for the high-throughput assessment of chemicals.
Data Quality and Chemical Space Coverage
A critical limitation of earlier benchmarks, particularly for open-source models, has been their reliance on relatively small datasets that may not adequately represent the chemical space of industrial drug discovery [22]. For instance, some widely used public solubility datasets contain only around 1,128 compounds, while PubChem holds over 14,000 relevant entries [22]. Furthermore, the mean molecular weight of compounds in some public benchmarks (e.g., 203.9 Dalton) is substantially lower than that of typical drug discovery compounds (300-800 Dalton), potentially limiting the real-world applicability of models trained on them [22].
To address this, recent initiatives like PharmaBench have employed large-scale data mining. This open-source benchmark was created using a multi-agent LLM (Large Language Model) system to identify and merge experimental conditions from 14,401 bioassays, resulting in a curated set of 52,482 entries across eleven ADMET datasets [22]. This represents a significant step forward in the scale and drug-likeness of open-source training data.
Commercial platforms, such as ADMET Predictor, often leverage premium datasets provided by pharmaceutical partners in addition to public data, which may enhance their performance on typical drug-like molecules [94].
Features and Usability
Beyond raw predictive accuracy, practical features and integration capabilities are key differentiators.
Table 3: Feature Comparison of Open-Source vs. Commercial ADMET Platforms
| Feature | Typical Open-Source Platforms | Typical Commercial Platforms (e.g., ADMET Predictor) |
|---|---|---|
| Core Capabilities | Prediction of fundamental ADMET properties [22]. | Prediction of >175 properties, including solubility vs. pH, logD curves, pKa, CYP metabolism, and toxicity [94]. |
| AI/ML Integration | Growing adoption of AI/ML models, as seen in PharmaBench's data mining [22]. | Flagship AI/ML platforms with integrated data analysis, SAR, and cheminformatics [94]. |
| Advanced Modeling | Basic QSAR and structure-based predictions. | Integrated high-throughput PBPK simulations and mechanistic safety assessments [94]. |
| Data & Applicability | Varies; newer benchmarks like PharmaBench offer improved chemical space coverage [22]. | Models trained on premium datasets; includes applicability domain and confidence estimates [94] [91]. |
| Workflow Integration | Script-based or standalone tools. | Enterprise-ready automation, REST APIs, Python wrappers, and integration with third-party platforms [94]. |
| Support & Updates | Community-driven support. | Professional customer support, documentation, and regular model updates [94]. |
Experimental Protocols for Validation
Protocol for Benchmarking Predictive Performance
Validating an ADMET platform requires a rigorous comparison of its predictions against reliable experimental data.
- Dataset Curation: Collect and curate external validation datasets from literature and public databases (e.g., ChEMBL, PubChem) [22] [91].
- Standardization: Standardize chemical structures (e.g., using RDKit), neutralize salts, and remove duplicates and inorganic compounds [91].
- Outlier Removal: Identify and remove intra-dataset outliers (e.g., using Z-score >3) and inter-dataset outliers (compounds with inconsistent values across sources) [91].
- Unit Consistency: Ensure all experimental values are converted to consistent units for the property being tested [91].
- Property Prediction: Use the software tools to generate in silico predictions for all compounds in the curated validation set(s).
- Performance Calculation: Calculate standard performance metrics for regression (e.g., R²) and classification (e.g., Balanced Accuracy) tasks [91].
- Applicability Domain Assessment: Evaluate whether the prediction chemicals fall within the model's applicability domain, as performance is typically more reliable for compounds within this domain [91].
Protocol for Integrating In Vitro Data
The following workflow outlines the process of using in silico predictions to guide experimental design and how in vitro results can, in turn, validate and refine the computational models.
Validation Feedback Workflow
The Scientist's Toolkit: Essential Research Reagents and Materials
The experimental validation of ADMET predictions relies on a suite of standardized in vitro assays and computational resources.
Table 4: Key Research Reagent Solutions for ADMET Validation
| Reagent / Material | Function in ADMET Validation |
|---|---|
| Caco-2 Cell Line | A model of the human intestinal barrier used to assess a compound's absorption potential via permeability assays [91] [95]. |
| Human Liver Microsomes | Contains cytochrome P450 enzymes; used to study a drug's metabolic stability and identify potential metabolites [95]. |
| Primary Hepatocytes | Fresh or cryopreserved human liver cells that provide a more complete metabolic system for studying hepatic clearance and toxicity [95]. |
| Assay Kits (Cytotoxicity) | Kits (e.g., MTT, LDH) used in high-throughput screening to evaluate compound-induced cellular toxicity [92]. |
| RDKit | An open-source cheminformatics toolkit used for chemical informatics, standardization of structures, and descriptor calculation in many open-source models [22] [91]. |
| Organ-on-a-Chip | Advanced microfluidic devices that emulate human organ physiology for more accurate absorption, metabolism, and toxicity testing [96] [95]. |
The choice between open-source and commercial ADMET platforms is not a matter of one being universally superior to the other. Instead, the decision should be guided by the specific needs, resources, and context of the research project. Open-source platforms have made remarkable strides in addressing data quality and scale issues, as evidenced by benchmarks like PharmaBench, making them highly valuable for academic research and groups with strong computational expertise [22]. Commercial platforms offer a compelling package of breadth, validated performance, enterprise-level support, and advanced integrated features like PBPK modeling that can accelerate decision-making in industrial drug discovery settings [94]. Ultimately, a robust ADMET prediction strategy should incorporate continuous validation against reliable in vitro data, regardless of the platform chosen, creating a feedback loop that strengthens both computational models and research outcomes.
The validation of in silico Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) predictions is a critical component of modern drug development. As the industry increasingly relies on New Approach Methodologies (NAMs) to prioritize compounds and reduce animal testing, establishing robust, quantitative frameworks for evaluating model performance has become essential [89]. Quantitative Structure-Property Relationship (QSPR) models provide a computational bridge between molecular structure and experimental outcomes, but their utility in regulatory and research decision-making depends entirely on rigorous, standardized validation against in vitro data [89] [97]. This guide objectively compares the key metrics and experimental protocols used to quantify QSPR model success, providing researchers with a standardized framework for model evaluation.
Core Metrics for QSPR Model Validation
Evaluating QSPR model performance requires a multi-faceted approach that captures different aspects of predictive accuracy and reliability. The following metrics are fundamental to this process.
Table 1: Key Quantitative Metrics for Evaluating QSPR Model Performance
| Metric | Formula/Description | Interpretation | Application Context |
|---|---|---|---|
| Root Mean Squared Log Error (RMSLE) | ( \sqrt{\frac{1}{n} \sum{i=1}^{n} (\log(yi + 1) - \log(\hat{y}_i + 1))^2 } ) | Measures relative error; less sensitive to large outliers. Ideal for widely varying data. | Predicting HTTK parameters like AUC and Css [89]. |
| Fold-Error Accuracy | Prediction within a specified fold-range (e.g., 1.5-fold, 2-fold) of experimental values. | An intuitive, binary measure of acceptable accuracy for a specific application. | QSAR-PBPK for fentanyl analogs (e.g., Vss error <1.5-fold deemed accurate) [97]. |
| Correlation Coefficients (R²/Rₚ) | ( R^2 = 1 - \frac{\sum (yi - \hat{y}i)^2}{\sum (y_i - \bar{y})^2} ) | Proportion of variance explained by the model. Rₚ is the Pearson correlation for predicted vs. actual. | Assessing relationship strength in QSPR models for physicochemical properties [98]. |
| Multiple Linear Regression (MLR) | ( y = b0 + b1x1 + b2x2 + ... + bnx_n + \epsilon ) | A statistical method to build and validate QSPR models, showing how descriptors jointly predict an activity. | Building QSAR models for biological activity (e.g., pIC₅₀) of antiviral drugs [98]. |
Experimental Protocols for Model Validation
Protocol 1: Validation with Time-Course In Vivo Data via PBPK Modeling
This advanced protocol uses in vivo data to indirectly assess the performance of QSPR-predicted in vitro parameters within a physiological context [89].
- Input Prediction: Utilize QSPR models to predict key in vitro parameters such as intrinsic hepatic clearance (Clint) and the fraction unbound in plasma (fup) [89].
- PBPK Simulation: Input the QSPR-predicted Clint and fup values into a high-throughput physiologically based pharmacokinetic (HT-PBTK) model.
- Output Simulation: Simulate in vivo plasma concentration-time profiles and derive toxicokinetic (TK) summary statistics, notably the area under the curve (AUC) and steady-state concentration (Css) [89].
- Performance Assessment: Compare the simulated TK summaries against actual in vivo time-course data (often from rats) using the RMSLE metric. A reported RMSLE of 0.6–0.8 for AUC using QSPR inputs indicates performance similar to models using in vitro measurements (RMSLE ~0.9) [89].
Protocol 2: Direct In Vitro to In Silico Comparison for PK Parameters
This method involves a direct head-to-head comparison of in silico predictions against in vitro experimental results for specific parameters [97].
- Data Generation: Obtain in vitro measured values for parameters such as tissue/blood partition coefficients (Kp).
- QSAR Prediction: Use a QSAR framework (e.g., the Lukacova method) to predict the same parameters based on chemical structure [97].
- Statistical Analysis: Calculate the fold-error between the QSAR-predicted values and the in vitro data. Predictions for volume of distribution at steady state (Vss) with an error of less than 1.5-fold, compared to errors exceeding 3-fold from interspecies extrapolation, validate the QSAR model's utility [97].
Protocol 3: QSPR/QSAR Model Construction and Validation for Physicochemical Properties
This protocol outlines the standard workflow for building and validating a QSPR model for properties like polarizability or biological activity like IC₅₀ [98].
- Descriptor Calculation: Represent chemical structures and compute molecular descriptors. Using molecular multigraphs (which account for double bonds) can yield higher correlation coefficients (e.g., R > 0.986 for polarizability) than simple graphs [98].
- Model Training: Employ Multiple Linear Regression (MLR) with a backward elimination approach (e.g., at a 0.05 significance level) to construct a model that relates the descriptors to the target property [98].
- Validation: Assess the model's predictive power by comparing its outputs against a test set of experimental data. High agreement, such as a predicted pIC₅₀ of 6.01 matching the observed value for Remdesivir, confirms model accuracy [98].
The following workflow diagram visualizes the key steps and decision points in a robust QSPR validation process, integrating the protocols described above.
The Scientist's Toolkit: Essential Research Reagents and Solutions
Successful QSPR validation relies on a combination of software tools, databases, and laboratory resources.
Table 2: Key Research Reagent Solutions for QSPR Validation
| Tool/Solution | Type | Primary Function in Validation |
|---|---|---|
| High-Throughput PBPK Model | Software/Algorithm | Simulates in vivo pharmacokinetics using in silico or in vitro inputs to enable indirect validation [89]. |
| CHEMBL Database | Bioactivity Database | Provides a large repository of experimentally validated bioactivity data (e.g., pIC₅₀, Ki) for training and benchmarking target-centric QSPR models [99]. |
| GastroPlus | PBPK Modeling Software | A commercial platform used for simulating and predicting pharmacokinetics, capable of integrating QSAR-predicted parameters like Kp [97]. |
| OPERA | QSPR Tool | Provides open-source, validated QSAR predictions for physicochemical properties, useful for generating input parameters or comparison data [89]. |
| Molecular Multigraph Tools | Computational Descriptor | Generates advanced molecular representations that can improve correlation in QSPR models for physicochemical properties compared to simple graphs [98]. |
The quantitative validation of QSPR models against in vitro data is a multi-dimensional process that extends beyond a single metric. A robust evaluation framework incorporates measures of relative error like RMSLE, intuitive thresholds like fold-error accuracy, and the statistical strength of correlation coefficients and MLR models. As demonstrated by case studies in toxicokinetics and fentanyl analog PK prediction, the choice of validation protocol—whether direct comparison, PBPK-enabled simulation, or full QSAR model construction—depends on the specific research question and data availability. By adhering to these standardized metrics and protocols, researchers can objectively compare model performance, build confidence in in silico predictions, and effectively integrate QSPR models into the drug development pipeline.
The field of toxicokinetics (TK), which describes the absorption, distribution, metabolism, and excretion (ADME) of chemicals by the body over time, is crucial for assessing chemical safety risks [89]. However, traditional methods for generating TK data face significant challenges. Animal studies, while informative, are resource-intensive, time-consuming, and raise ethical concerns, making them impractical for evaluating the vast number of environmental chemicals lacking data [89]. Regulatory frameworks like REACH and TSCA have further increased the need for efficient testing approaches by calling for reductions in vertebrate testing [89].
High-Throughput Toxicokinetic (HTTK) approaches address this data gap by combining efficiently obtained chemical-specific data with chemical-agnostic mathematical models [89]. These methods enable risk-based prioritization of thousands of chemicals by predicting key TK parameters such as area under the curve (AUC) and steady-state concentration (Css) [89]. HTTK traditionally relies on in vitro measurements for parameters like intrinsic hepatic clearance (Clint) and fraction unbound in plasma (fup) [89]. While in vitro methods have advanced significantly, generating these data for thousands of chemicals remains costly and time-consuming, with data available for only approximately 1,000 chemicals to date [89] [100].
In silico predictions using Quantitative Structure-Property Relationship (QSPR) models offer a promising alternative by estimating HTTK parameters directly from chemical structure [89]. Multiple QSPR models have been developed to predict key parameters including Clint, fup, and elimination half-life (t½), ranging from open-source models based on public data to proprietary models using large proprietary datasets [89]. As noted in a recent LinkedIn post by researcher John Wambaugh, "QSPR models perform comparably to in vitro data, with mean predictions within a 16-fold range" [100]. This case study examines a collaborative evaluation of seven QSPR models from six international groups, assessing their performance against both in vitro measurements and in vivo data through physiologically based TK (PBTK) modeling [89].
Methodology
Collaborative Framework and Participating Models
This collaborative evaluation involved seven QSPR models from six international modeling groups representing academic, regulatory, and commercial entities [89]. The study was designed to overcome the challenges of comparing QSPR models built with different training data, optimization metrics, and chemical domains [89]. Participants were provided with chemical identities and structure descriptors but were blinded to the in vivo evaluation data to prevent bias [89].
Table: QSPR Models Evaluated in the Collaborative Study
| Modeled Parameter | Number of Models | Model Characteristics |
|---|---|---|
| Intrinsic Hepatic Clearance (Clint) | Multiple models | Predicts hepatic metabolic clearance using chemical structure |
| Fraction Unbound in Plasma (fup) | Multiple models | Estimates plasma protein binding based on molecular properties |
| TK Elimination Half-Life (t½) | 2 models | Directly predicts in vivo elimination half-life |
The evaluation framework employed three levels of analysis, progressing from direct parameter comparison to whole-body TK prediction [89]:
- Level 1: Direct comparison of QSPR-predicted HTTK parameters against in vitro measured values for chemicals where both were available
- Level 2: Evaluation using in vivo TK data, mainly from rats, with simulations performed using a high-throughput PBTK model
- Level 3: Sensitivity analysis to determine how Clint and fup parameters inform predictions of AUC and Css
QSPR Model Predictions
Modeling groups generated predictions for Clint, fup, and/or t½ using their respective QSPR approaches [89]. The models ranged from freely available, open-source implementations to proprietary systems underpinned by large datasets [89]. Some models followed Organisation for Economic Co-operation and Development (OECD) validation principles for regulatory application [89].
In Vitro Data
The study used existing in vitro HTTK data for parameters including Clint and fup, obtained through high-throughput in vitro assays [89]. These measurements served as the reference for evaluating QSPR model performance at Level 1 of the analysis.
In Vivo Toxicokinetic Data
The evaluation utilized literature time-course in vivo TK data, primarily from rats, to assess the real-world predictive capability of the PBTK models parameterized with both in vitro and QSPR-predicted values [89]. This approach allowed researchers to estimate how well the combined QSPR-PBTK framework could predict actual concentration-time profiles observed in living organisms.
Physiologically Based Toxicokinetic (PBTK) Modeling
A critical component of the methodology involved implementing a high-throughput PBTK (HT-PBTK) model to simulate in vivo plasma concentration-time profiles [89]. This model was parameterized using two distinct approaches:
- In vitro approach: Using experimentally measured Clint and fup values as inputs
- QSPR approach: Using QSPR-predicted Clint and fup values as inputs
The performance of both approaches was evaluated by comparing their simulations against the actual in vivo time-course data [89]. The root mean squared log10 error (RMSLE) metric was used to quantify predictive accuracy for key TK metrics including AUC and Css [89].
Diagram Title: HTTK QSPR Evaluation Workflow
Research Reagent Solutions
Table: Essential Research Reagents and Computational Tools for HTTK Evaluation
| Category | Specific Tool/Reagent | Function in Evaluation |
|---|---|---|
| In Vitro Assays | Hepatic clearance assays | Measurement of intrinsic hepatic clearance (Clint) |
| Plasma protein binding assays | Determination of fraction unbound in plasma (fup) | |
| In Vivo Data | Rat toxicokinetic studies | Time-course plasma concentration data for validation |
| Computational Tools | OPERA | Predicts physicochemical properties from structure [89] |
| QSPR models | Predicts HTTK parameters (Clint, fup, t½) from structure [89] | |
| High-throughput PBTK model | Simulates plasma concentration-time profiles [89] | |
| Data Analysis | RMSLE calculation | Quantifies prediction error for TK parameters [89] |
| Sensitivity analysis | Determines influence of Clint/fup on AUC and Css [89] |
Results and Discussion
Performance Comparison: QSPR Predictions vs. In Vitro Measurements
The Level 1 evaluation provided critical insights into the agreement between QSPR-predicted parameters and traditional in vitro measurements. For a subset of chemicals with available in vitro data, QSPR models demonstrated the capability to predict Clint and fup within a reasonable error range [89]. The collaborative nature of the study revealed that different QSPR approaches showed varying performance depending on the chemical space and specific parameter being predicted [89].
Notably, the study estimated that using rat in vivo data to evaluate QSPR models trained on human in vitro data might inflate error estimates by as much as RMSLE 0.8, highlighting the challenges of cross-species extrapolation in TK prediction [89]. This finding underscores the importance of consistent training and evaluation datasets when assessing model performance.
Toxicokinetic Prediction Accuracy
The core of the evaluation compared the ability of PBTK models parameterized with either in vitro measurements or QSPR predictions to simulate in vivo TK profiles. The comprehensive assessment yielded the following key findings regarding prediction accuracy for critical TK metrics:
Table: Toxicokinetic Prediction Performance Metrics
| Parameter Source | TK Metric | Prediction Performance (RMSLE) | Comparative Assessment |
|---|---|---|---|
| In Vitro Measurements | AUC | 0.9 | Baseline performance using traditional HTTK approach |
| QSPR Predictions | AUC | 0.6 - 0.8 | Outperforms in vitro parameter approach |
| In Vitro Measurements | Css | Similar to AUC trends | Established reference for Css prediction |
| QSPR Predictions | Css | Similar to AUC trends | Comparable or superior to in vitro approach |
| Both Approaches | Overall TK | PBTK with QSPR performed similarly to using in vitro values (RMSLE ∼1) | QSPR-based approach provides viable alternative |
The sensitivity analysis confirmed that both Clint and fup parameters significantly influence predictions of AUC and Css, explaining why accurate estimation of these parameters is crucial for reliable TK prediction [89]. As researcher John Wambaugh noted in a LinkedIn post summarizing the findings, "For chemicals suitable for HTTK, QSPRs offer a viable alternative when in vitro data are unavailable" [100].
Diagram Title: TK Prediction Performance Results
Implications for In Silico ADMET Prediction Validation
This case study provides significant insights for the broader validation of in silico ADMET predictions, particularly in the context of regulatory acceptance and practical application in drug discovery and chemical safety assessment.
Addressing Data Quality and Consistency Challenges
The findings align with recent research highlighting critical challenges in molecular property prediction, including data heterogeneity and distributional misalignments that can compromise predictive accuracy [101]. Significant misalignments have been identified between gold-standard and popular benchmark sources for ADME properties, with dataset discrepancies arising from differences in experimental conditions and chemical space coverage [101]. These inconsistencies can introduce noise and ultimately degrade model performance, underscoring the importance of rigorous data consistency assessment prior to modeling [101].
Tools like AssayInspector have been developed to systematically characterize datasets by detecting distributional differences, outliers, and batch effects that could impact machine learning model performance [101]. This aligns with the collaborative approach taken in the HTTK case study, where multiple models were evaluated against standardized datasets to ensure robust performance assessment.
Regulatory and Practical Implementation
From a regulatory perspective, the demonstration that QSPR-based approaches can perform similarly to in vitro-based methods supports their use in screening and priority-setting exercises. As noted in the study, "We anticipate that, for some novel compounds, QSPRs for HTTK input parameters will give predictions of TK similar to those based on in vitro measurements" [89]. This is particularly valuable for regulatory programs that require TK information for large numbers of chemicals but face practical constraints on in vitro testing capacity.
The case study also highlights the potential of federated learning approaches for enhancing ADMET prediction models. Recent advances demonstrate that federation "alters the geometry of chemical space a model can learn from, improving coverage and reducing discontinuities in the learned representation" [9]. Federated models have been shown to systematically outperform local baselines, with performance improvements scaling with the number and diversity of participants [9]. This approach could address the limitation observed in the HTTK evaluation where model performance varied across chemical space.
This collaborative evaluation demonstrates that QSPR models for predicting HTTK parameters can provide TK predictions comparable to those based on in vitro measurements, with PBTK models using QSPR predictions performing similarly to those using in vitro values (RMSLE ∼1) [89]. The finding that AUC can be predicted with RMSLE of 0.6-0.8 using QSPR model values, potentially outperforming predictions based on in vitro measurements (RMSLE 0.9), supports the use of QSPR approaches as viable alternatives when in vitro data are unavailable [89].
These results have significant implications for advancing new approach methodologies (NAMs) in chemical safety assessment and drug discovery. By providing a validated framework for in silico toxicokinetic prediction, this approach supports the reduction of animal testing while expanding coverage to the thousands of chemicals lacking experimental data. The collaborative nature of the study, involving multiple international groups across academic, regulatory, and commercial sectors, enhances confidence in the conclusions and provides a model for future evaluations of in silico prediction methods.
As the field progresses, addressing challenges related to data quality, model interpretability, and domain of applicability will be essential for regulatory acceptance and broader implementation. The integration of advanced approaches such as federated learning and rigorous data consistency assessment will further enhance the reliability and applicability of in silico HTTK predictions for chemical safety assessment and drug discovery.
For researchers and drug development professionals, gaining regulatory approval from the Food and Drug Administration (FDA) and European Medicines Agency (EMA) requires demonstrating that your in silico ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) predictions are scientifically valid and reliably predict clinical outcomes. Regulatory submissions are comprehensive packages of documents and data submitted to health authorities to demonstrate the safety, efficacy, and quality of a drug [102]. With promising drug candidates often failing due to suboptimal ADME properties or toxicity concerns, the validation of predictive models has become crucial for building regulatory confidence and improving the efficiency of drug development [1] [33].
This guide provides a structured approach for preparing validation dossiers that meet the distinct but overlapping expectations of both FDA and EMA, particularly within the context of validating in silico ADMET models with experimental in vitro data.
Regulatory Landscape: Comparing FDA and EMA Requirements
While both agencies accept the Common Technical Document (CTD) format and share the fundamental goal of ensuring product quality, safety, and efficacy, differences exist in their detailed requirements and philosophical approaches to process validation [102] [103].
Table 1: Key Regulatory Differences Between FDA and EMA
| Aspect | FDA (U.S. Food and Drug Administration) | EMA (European Medicines Agency) |
|---|---|---|
| Application Type | New Drug Application (NDA) / Biologics License Application (BLA) [103] | Marketing Authorisation Application (MAA) [103] |
| Primary Committee | CDER (drugs) / CBER (biologics) [103] | CHMP (drugs) / CAT (advanced therapies) [103] |
| Process Validation Lifecycle | Clearly defined three-stage model (Process Design, Process Qualification, Continued Process Verification) [104] | Life-cycle focused, covering prospective, concurrent and retrospective validation [104] |
| Validation Master Plan (VMP) | Not mandatory, but expects an equivalent structured document [104] | Mandatory [104] |
| Ongoing Verification | Continued Process Verification (CPV), data-driven with high emphasis on statistics [104] | Ongoing Process Verification (OPV), based on real-time or retrospective data, incorporated in Product Quality Review [104] |
| Batch Requirements | Minimum of three commercial batches recommended for Process Qualification [104] | Risk-based, requires scientific justification rather than a mandated number [104] |
The path to approval involves rigorous evaluation. The EMA's evaluation timeline is typically 210 days, plus up to two "clock stops" for applicants to respond to questions [103]. The FDA aims to review standard NDAs within 10 months and priority applications in 6 months [103]. Both agencies offer opportunities for interaction, such as the FDA's Type A, B, and C meetings, and the EMA's Scientific Advice procedures, which can be conducted in parallel (Parallel Scientific Advice) to align development strategies [103].
Validating In Silico ADMET Models: A Framework for Regulatory Submissions
The core of building regulatory confidence lies in providing robust, scientifically sound evidence that your computational models are predictive of human outcomes. Machine Learning (ML) and Artificial Intelligence (AI) have revolutionized ADMET prediction by deciphering complex structure-property relationships, offering scalable and efficient alternatives to traditional experimental methods [33]. However, their "black box" nature can impede interpretability, posing a challenge for regulatory acceptance [33].
A successful validation dossier must bridge the gap between in silico predictions and empirical data, following a logical workflow.
Figure 1: The Validation Workflow from model development to regulatory submission.
Experimental Design for In Vitro Validation
To validate in silico ADMET predictions, you must generate high-quality, physiologically relevant in vitro data. The following table outlines key experiments and their protocols that serve as the cornerstone of a robust validation package.
Table 2: Key Experimental Protocols for ADMET Validation
| ADMET Property | Common In Vitro Assay | Detailed Methodology & Endpoint Measurements |
|---|---|---|
| Absorption / Permeability | Caco-2 Permeability [105] [106] | - Cultivate Caco-2 cells on semi-permeable membranes for 21+ days to form differentiated monolayers.- Measure Trans-Epithelial Electrical Resistance (TEER) to confirm monolayer integrity.- Apply test compound to the apical (donor) compartment and sample from the basolateral (receiver) compartment over time.- Analyze samples using LC-MS to determine apparent permeability (Papp) and Efflux Ratio [105]. |
| Metabolism / Clearance | Hepatic Metabolic Stability [106] [107] | - Incubate test compound with human liver microsomes or highly functional, metabolically competent hepatocytes in long-term culture.- Collect samples at multiple time points (e.g., 0, 15, 30, 60, 90 minutes).- Terminate reactions with an organic solvent (e.g., acetonitrile).- Analyze parent compound disappearance via LC-MS to calculate intrinsic clearance (CL~int~) and identify metabolites [107]. |
| Drug-Drug Interaction (Metabolism) | Cytochrome P450 (CYP) Inhibition [1] | - Incubate human liver microsomes with a CYP-specific probe substrate (e.g., testosterone for CYP3A4) in the presence and absence of the test compound.- Measure the formation of the specific metabolite using LC-MS/MS.- Calculate the percentage inhibition and IC~50~ values to assess the compound's potential to cause drug-drug interactions [1]. |
| Bioavailability Prediction | Multi-Organ Model (e.g., Gut-Liver) [107] | - Utilize connected gut and liver in vitro models to simulate first-pass metabolism.- Apply compound to the gut compartment and measure its passage and metabolism before reaching the liver compartment.- Sample from the "systemic circulation" compartment.- Use mechanistic mathematical modeling of the experimental data to predict human oral bioavailability (F) [107]. |
The Scientist's Toolkit: Essential Research Reagents and Solutions
The reliability of your validation data depends heavily on the quality and consistency of the materials used. The table below details key reagents and their critical functions in ADMET assay workflows.
Table 3: Essential Research Reagent Solutions for ADMET Assays
| Reagent / Solution | Function in Experimental Protocol |
|---|---|
| Caco-2 Cell Line | A human colon adenocarcinoma cell line that, upon differentiation, forms a polarized monolayer with tight junctions and expresses relevant transporters (e.g., P-gp). It is the gold standard model for predicting intestinal permeability [105]. |
| Cryopreserved Human Hepatocytes | Metabolically competent cells that contain a full complement of drug-metabolizing enzymes and transporters. They are essential for generating human-relevant data on metabolic stability, metabolite identification, and enzyme induction/inhibition [107]. |
| Human Liver Microsomes (HLM) | Subcellular fractions containing membrane-bound cytochrome P450 (CYP) and other enzymes. A cost-effective and standardized system for high-throughput assessment of phase I metabolic stability and CYP inhibition potential [1] [106]. |
| Transwell Permeable Supports | Multi-well plates with membrane inserts that allow for the growth of cell monolayers and separate apical and basolateral compartments. Critical for permeability (Caco-2) and transport studies [105]. |
| Protein-Free Cell Culture Medium (Specialized) | Used in conjunction with low non-specific binding assay plates to accurately quantify drug concentrations and parameters like protein binding and intrinsic clearance without interference from medium components [107]. |
| LC-MS/MS Grade Solvents | High-purity solvents (e.g., water, acetonitrile, methanol) essential for mobile phase preparation and sample processing in Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) to ensure sensitive and unambiguous analyte detection [105] [107]. |
Building the Validation Dossier: From Data to CTD
The final step is to compile all evidence into a compelling validation dossier structured according to the CTD format. The dossier should tell a coherent story about the predictive power and robustness of your in silico models.
Data Integration and Correlation Analysis
The heart of your dossier is the demonstration of a strong correlation between your in silico predictions and the experimentally derived in vitro data. This can be visualized as follows:
Figure 2: The core logic of model validation, linking prediction and experiment.
For the correlation analysis, present quantitative data clearly. The table below provides a hypothetical example of how to structure such results for a Caco-2 permeability model.
Table 4: Example Data Table for Caco-2 Permeability Model Validation
| Compound ID | In Silico Prediction (Papp x10⁻⁶ cm/s) | In Vitro Experimental Result (Papp x10⁻⁶ cm/s) | Residual (Predicted - Experimental) | Clinical Permeability Classification |
|---|---|---|---|---|
| CPD-001 | 12.5 | 15.2 | -2.7 | High |
| CPD-002 | 5.2 | 4.8 | +0.4 | Moderate |
| CPD-003 | 1.1 | 0.9 | +0.2 | Low |
| CPD-004 | 22.3 | 18.7 | +3.6 | High |
| CPD-005 | 3.8 | 5.1 | -1.3 | Moderate |
Summary Statistics: R² = 0.94, Root Mean Square Error (RMSE) = 2.1 x10⁻⁶ cm/s, Mean Absolute Error (MAE) = 1.6 x10⁻⁶ cm/s.
Addressing Specific Regulatory Expectations in the Dossier
- For the FDA: Emphasize the data-driven, statistical rigor of your validation approach, aligning with the spirit of Continued Process Verification (CPV) [104]. Clearly document the model's performance using robust statistical metrics and demonstrate its application across the product lifecycle, from lead optimization to clinical candidate selection.
- For the EMA: Explicitly reference and fulfill the requirements of Annex 15 of the EU GMP Guidelines [104]. Include a Validation Master Plan (VMP) for your computational models that outlines the scope, responsibilities, and timelines of the validation activities. Provide a strong scientific justification for your validation approach, including the number and diversity of compounds used in the validation set [104].
Successfully navigating the regulatory landscape for FDA and EMA submissions requires a strategic and evidence-based approach to validating in silico ADMET tools. By understanding the nuanced expectations of each agency, implementing robust and biorelevant in vitro experimental protocols, and systematically correlating this data with computational predictions, you can build a powerful validation dossier. This dossier, structured within the CTD framework, will provide the scientific evidence needed to build regulatory confidence, de-risk drug development, and accelerate the journey of safer, more effective therapeutics to patients.
Conclusion
The successful integration of in silico ADMET predictions with in vitro validation is no longer optional but a fundamental component of efficient and predictive drug discovery. This synthesis demonstrates that a multi-faceted approach—combining advanced AI models, high-quality curated datasets, physiologically relevant in vitro systems, and rigorous validation frameworks—is essential for bridging the translational gap. Future progress will be driven by enhancing model interpretability, expanding the use of human-relevant advanced in vitro models like organ-on-a-chip to generate high-quality data, and fostering closer collaboration between industry, academia, and regulators. By adopting these integrated strategies, researchers can significantly de-risk drug development, improve candidate selection, and accelerate the delivery of safer, more effective therapeutics to patients.
References
- 1. In Silico ADME Methods Used in the Evaluation of Natural ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12389637/]
- 2. ADMET | 药物分子性质智能化评估平台,助力创新药物研发 [https://ins.sjtu.edu.cn/people/lhong/news/news10.html]
- 3. Nature Medicine综述:AI药物研发最新进展原创 [https://blog.csdn.net/weixin_58753619/article/details/147618873]
- 4. ADMET Predictor 软件的ADMET/PK性质预测模块介绍与操作演示 [https://www.pharmogo.com/?news_15/773.html]
- 5. 连接数据与药物开发:用于下一代ADMET预测的机器学习方法 [http://www.360doc.com/content/25/1030/08/71609523_1164002707.shtml]
- 6. 又双叒叕升级了!ADMETlab 3.0——全面升级的药物 ... [https://hub.baai.ac.cn/view/36325]
- 7. 开源软件ADMET-AI的介绍 [http://blog.molcalx.com.cn/2024/12/05/admet-ai.html]
- 8. 小分子化合物人体药代动力学参数预测的方法及应用 [https://dmpkservice.wuxiapptec.com/cn/blogs/456-id/]
- 9. Federated Learning for ADMET Prediction [https://www.apheris.com/resources/blog/federated-learning-for-admet-prediction-expanding-model-applicability]
- 10. In silico ADMET prediction: recent advances, current ... [https://pubmed.ncbi.nlm.nih.gov/23675935/]
- 11. Revolutionizing pharmacology: AI-powered approaches in ... [https://www.sciencedirect.com/science/article/pii/S259009862500020X]
- 12. ADMET evaluation in drug discovery: 21. Application and ... [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-025-00947-z]
- 13. Comparative Analysis of Chemical Distribution Models for ... [https://www.mdpi.com/2305-6304/13/6/439]
- 14. Benchmarking ML in ADMET predictions: the practical impact ... [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-025-01041-0]
- 15. Predicting ADMET Properties from Molecule SMILE [https://pmc.ncbi.nlm.nih.gov/articles/PMC11207804/]
- 16. Using ADMET to Move Forward from Drug Discovery ... [https://bitesizebio.com/24006/using-admet-to-move-forward-from-drug-discovery-to-development/]
- 17. Enhancing drug screening and pharmacokinetic predictions [https://www.sciencedirect.com/science/article/abs/pii/S1385894725012288]
- 18. Evaluation of Free Online ADMET Tools for Academic or ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9864198/]
- 19. ADMET evaluation in drug discovery: 21. Application and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11724520/]
- 20. The Trends and Future Prospective of In Silico Models from ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10675155/]
- 21. Characterization, in-silico, and in-vitro study of a new ... [https://www.nature.com/articles/s41598-022-09809-2]
- 22. PharmaBench: Enhancing ADMET benchmarks with large ... [https://www.nature.com/articles/s41597-024-03793-0]
- 23. In Search of Beautiful Molecules: A Perspective on Generative ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12458704/]
- 24. ICH M12 Guideline Overview on Drug Interaction Studies [https://www.dlrcgroup.com/ich-m12-guideline-overview-on-drug-interaction-studies/]
- 25. The New Drug-Drug Interaction Guidance, ICH M12 [https://premier-research.com/perspectives/the-new-drug-drug-interaction-guidance-ich-m12-how-to-accelerate-your-drug-development-program/]
- 26. The New ICH M12 Guideline – Harmony at Last for DDI ... [https://www.evotec.com/sciencepool/the-new-ich-m12-guideline-harmony-at-last-for-ddi-studies]
- 27. M12 Drug Interaction Studies August 2024 [https://www.fda.gov/regulatory-information/search-fda-guidance-documents/m12-drug-interaction-studies]
- 28. ICH M12 on drug interaction studies - Scientific guideline [https://www.ema.europa.eu/en/ich-m12-drug-interaction-studies-scientific-guideline]
- 29. New harmonized ICH M12 guideline for drug interaction ... [https://admescope.com/welcoming-the-new-harmonized-ich-m12-guideline-for-drug-interaction-studies/]
- 30. ICH M12 Guidelines & Your Drug-Drug Interaction Package [https://www.certara.com/blog/ich-m12-guidelines-and-your-drug-drug-interaction-package/]
- 31. ICH M12 Drug Interaction Studies: Summary of the Efforts ... [https://pubmed.ncbi.nlm.nih.gov/40108838/]
- 32. Open access in silico tools to predict the ADMET profiling ... [https://pubmed.ncbi.nlm.nih.gov/32735147/]
- 33. Machine learning approaches for next-generation ADMET ... [https://www.sciencedirect.com/science/article/abs/pii/S1359644625002004]
- 34. Bayer's in silico ADMET platform: a journey of machine ... [https://www.sciencedirect.com/science/article/pii/S1359644620302609]
- 35. Differences between in vitro, in vivo, and in silico studies [https://mpkb.org/home/patients/assessing_literature/in_vitro_studies]
- 36. In Vitro vs. In Vivo vs. In Silico: Preclinical Assay Differences [https://www.zeclinics.com/blog/differences-between-in-vitro-in-vivo-and-in-silico-assays-in-preclinical-research]
- 37. Development and validation of machine learning models ... [https://www.nature.com/articles/s41598-025-06998-4]
- 38. Embryo selection with artificial intelligence: how to evaluate ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8324599/]
- 39. Comprehensive benchmarking of computational tools for ... [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-024-00931-z]
- 40. In Silico ADME Methods Used in the Evaluation of Natural ... [https://www.mdpi.com/1999-4923/17/8/1002]
- 41. In silico and ADMET molecular analysis targeted to ... [https://www.sciencedirect.com/science/article/pii/S2405580824001225]
- 42. A Combination of Machine Learning and PBPK Modeling ... [https://link.springer.com/article/10.1007/s11095-024-03725-y]
- 43. Opportunities for machine learning and artificial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12573771/]
- 44. Applications of PBPK Modeling to Estimate Drug Metabolism ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12473325/]
- 45. Prediction of pharmacokinetic/pharmacodynamic ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1578117/full]
- 46. Advanced ADMET Modeling Using Deep Learning and AI [https://www.technologynetworks.com/drug-discovery/blog/beyond-black-box-models-advances-in-data-driven-admet-modeling-403114]
- 47. Toward in silico structure-based ADMET prediction in drug ... [https://www.sciencedirect.com/science/article/abs/pii/S135964461100362X]
- 48. Comparative Analysis of Chemical Distribution Models for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12196727/]
- 49. ADMET Benchmark Group [https://www.emergentmind.com/topics/admet-benchmark-group]
- 50. Permeability Benchmarking: Guidelines for Comparing in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11815851/]
- 51. PBPK modeling and simulation in drug research ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5125732/]
- 52. The Evolution and Future Directions of PBPK Modeling in ... [https://www.mdpi.com/1999-4923/17/11/1413]
- 53. Integrating PBPK models with cell-based assays [https://www.sciencedirect.com/science/article/pii/S0041008X2500211X]
- 54. In silico trials: Verification, validation and uncertainty ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7883933/]
- 55. Time For a New Adventure - Practical Cheminformatics [https://patwalters.github.io/Time-For-a-New-Adventure/]
- 56. Virtual screening, ADMET prediction, molecular docking ... [https://www.nature.com/articles/s41598-024-75292-6]
- 57. In Silico exploration of Karanjin for anti-obesity potential ... [https://www.nature.com/articles/s41598-025-14257-9]
- 58. In silico evaluation of pharmacokinetic properties and ... [https://www.sciencedirect.com/science/article/pii/S1476927125002877]
- 59. Identification of natural compounds as potential inhibitors ... [https://www.frontiersin.org/journals/drug-discovery/articles/10.3389/fddsv.2025.1525533/full]
- 60. Accelerating AI Interpretability [https://fas.org/publication/accelerating-ai-interpretability/]
- 61. Top 10 Tools for Achieving AI Transparency and ... [https://superagi.com/top-10-tools-for-achieving-ai-transparency-and-explainability-in-2025-2/]
- 62. Synthesis, in silico, and in vitro pharmacological evaluation ... [https://japsonline.com/abstract.php?article_id=4540&sts=2]
- 63. Mastering Explainable AI in 2025: A Beginner's Guide to ... [https://superagi.com/mastering-explainable-ai-in-2025-a-beginners-guide-to-transparent-and-interpretable-models/]
- 64. The Challenge of AI Interpretability [https://www.deloitte.com/us/en/what-we-do/capabilities/applied-artificial-intelligence/articles/ai-interpretability-challenge.html]
- 65. Leveraging machine learning models in evaluating ADMET ... [https://pubmed.ncbi.nlm.nih.gov/40585410/]
- 66. Uncertainties in the Extrapolation of In Vitro Data in Human ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12175163/]
- 67. Challenges and Opportunities for In Vitro–In Vivo ... [https://www.sciencedirect.com/science/article/pii/S0090955624010973]
- 68. Early-Stage PROTAC Development and the Road to IND [https://www.aapsnewsmagazine.org/cover-story-oct25]
- 69. The Future of PROTAC Research: Emerging Opportunities ... [https://panomebio.com/blog/the-future-of-protac-research-emerging-opportunities-and-challenges/]
- 70. Overcoming ADME Challenges in PROTAC Development [https://www.syngeneintl.com/resources/blog/overcoming-adme-challenges-in-protac-development-key-insights-from-syngene-experts/]
- 71. Optimising proteolysis-targeting chimeras (PROTACs) for ... [https://www.sciencedirect.com/science/article/abs/pii/S1359644620302932]
- 72. In Vitro ADME Profiling of PROTACs [https://pubmed.ncbi.nlm.nih.gov/40273026/]
- 73. Recent Advances in the Development of Pro-PROTAC for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12473374/]
- 74. A primary human Gut/Liver microphysiological system to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12597587/]
- 75. Publication | Gut/Liver MPS | Human Oral Bioavailability [https://cn-bio.com/resource/a-primary-human-gut-liver-microphysiological-system-to-estimate-human-oral-bioavailability/]
- 76. Blog | In vitro human Gut/Liver models for ADME [https://cn-bio.com/in-vitro-human-gut-liver-models/]
- 77. Oral bioavailability property prediction based on task ... [https://pubmed.ncbi.nlm.nih.gov/40928678/]
- 78. QSAR Models for Predicting Oral Bioavailability and ... [https://www.mdpi.com/2039-4713/15/5/166]
- 79. Artificial intelligence-enhanced bioavailability studies [https://www.sciencedirect.com/science/article/abs/pii/S0924224425003395]
- 80. A Comprehensive Review of Organ - on - a - Chip Technology and Its... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11118005/]
- 81. Organ-on-a-chip adoption: The roadmap to broader use [https://cn-bio.com/organ-on-a-chip-adoption-the-roadmap-to-broader-use/]
- 82. In silico modelling of organ-on-a-chip devices: an overview [https://www.frontiersin.org/journals/bioengineering-and-biotechnology/articles/10.3389/fbioe.2024.1520795/full]
- 83. Using Organ-on-a-Chip Technology to Unlock Patient ... [https://emulatebio.com/using-organ-on-a-chip-technology-to-unlock-patient-derived-precision-medicine/]
- 84. Blog | Integrating in silico tools with Organ-on-a-chip [https://cn-bio.com/integrating-in-silico-tools-with-organ-on-a-chip-to-advance-adme-studies/]
- 85. Latest advancements in organ-on-a-chip technology - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12163327/]
- 86. - Organ - on - a : the next generation platform for risk assessment of... chip [https://pubs.rsc.org/en/content/articlehtml/2020/ra/d0ra05173j]
- 87. Benchmarking ML in ADMET predictions: the practical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12281724/]
- 88. DeepDelta: predicting ADMET improvements of molecular ... [https://jcheminf.biomedcentral.com/articles/10.1186/s13321-023-00769-x]
- 89. Collaborative evaluation of in silico predictions for high ... [https://www.sciencedirect.com/science/article/abs/pii/S0887233325001444]
- 90. ADMET Predictor Simulations in Drug Discovery Screening [https://www.pharmaron.com/knowledge-center/admet-predictor-simulations/]
- 91. Comprehensive benchmarking of computational tools for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11674477/]
- 92. Top Market Shifts Transforming the Pharma ADMET ... [https://www.openpr.com/news/4272440/top-market-shifts-transforming-the-pharma-admet-testing-market]
- 93. Enhancing preclinical drug discovery with artificial ... [https://www.sciencedirect.com/science/article/pii/S1359644621005043]
- 94. ADMET Predictor® Modules [https://www.simulations-plus.com/software/admetpredictor/]
- 95. In Vitro ADME-Tox Market Regional Trends, Growth & ... [https://www.linkedin.com/pulse/vitro-adme-tox-market-regional-trends-growth-share-oobge/]
- 96. In Vitro ADME 2025 Trends and Forecasts 2033 [https://www.archivemarketresearch.com/reports/in-vitro-adme-139485]
- 97. model validation, human pharmacokinetic prediction and ... [https://pubmed.ncbi.nlm.nih.gov/41181589/]
- 98. QSPR/QSAR study of antiviral drugs modeled as ... [https://www.nature.com/articles/s41598-024-63007-w]
- 99. A precise comparison of molecular target prediction methods [https://pubs.rsc.org/en/content/articlehtml/2025/dd/d5dd00199d]
- 100. QSPR models predict HTTK in vitro parameters [https://www.linkedin.com/posts/john-wambaugh-90a2586_collaborative-evaluation-of-in-silico-predictions-activity-7379563218823696384-cx33]
- 101. Enhancing molecular property prediction through data ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12574261/]
- 102. A Guide to Regulatory Submissions in the US (FDA) & EU ... [https://kivo.io/news/a-regulatory-submissions-guide-for-the-us-fda-eu-ema]
- 103. EMA Marketing Authorisation vs FDA New Drug Application [https://somerville-partners.com/marketing-authorisation-application-ema-vs-new-drug-application-fda/]
- 104. EMA vs FDA Expectations on Process Validation [https://www.pharmaguideline.com/2025/07/ema-vs-fda-expectations-on-process-validation.html]
- 105. ADMET evaluation in drug discovery: 21. Application and ... [https://pubmed.ncbi.nlm.nih.gov/39794857/]
- 106. Computational approaches to DMPK: A realistic ... [https://www.sciencedirect.com/science/article/pii/S1359644625001357]
- 107. In Vitro ADME Services | Organ-on-a-Chip Studies [https://cn-bio.com/in-vitro-services/adme/]